Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
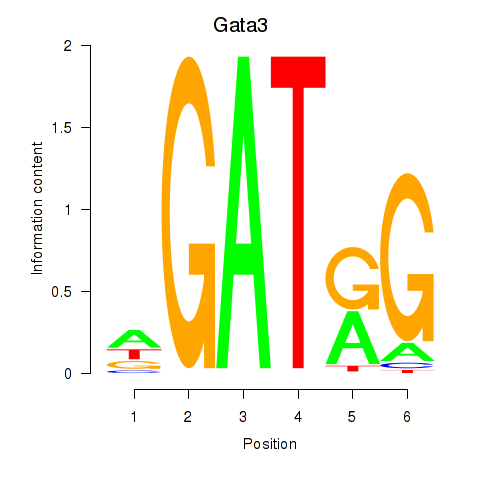
Results for Gata3
Z-value: 2.31
Transcription factors associated with Gata3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata3
|
ENSMUSG00000015619.11 | Gata3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata3 | mm39_v1_chr2_-_9883391_9883418 | 0.08 | 5.1e-01 | Click! |
Activity profile of Gata3 motif
Sorted Z-values of Gata3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_95426419 | 17.91 |
ENSMUST00000229933.2
ENSMUST00000166170.9 |
Nell2
|
NEL-like 2 |
| chr11_+_58839716 | 16.26 |
ENSMUST00000078267.5
|
H2bu2
|
H2B.U histone 2 |
| chr13_+_46655617 | 12.40 |
ENSMUST00000225824.2
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr11_+_87651359 | 11.96 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr6_-_58884038 | 10.84 |
ENSMUST00000059539.5
|
Nap1l5
|
nucleosome assembly protein 1-like 5 |
| chr4_+_127066667 | 10.68 |
ENSMUST00000106094.9
|
Dlgap3
|
DLG associated protein 3 |
| chr9_+_107822458 | 10.28 |
ENSMUST00000194206.2
|
Camkv
|
CaM kinase-like vesicle-associated |
| chr13_+_46655324 | 10.26 |
ENSMUST00000021802.16
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr16_-_23709564 | 10.23 |
ENSMUST00000004480.5
|
Sst
|
somatostatin |
| chr1_-_132295617 | 10.02 |
ENSMUST00000142609.8
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr3_+_94386385 | 9.95 |
ENSMUST00000199775.5
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr17_+_35454833 | 9.41 |
ENSMUST00000118384.8
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr5_-_121148143 | 9.39 |
ENSMUST00000202406.4
ENSMUST00000200792.2 |
Rph3a
|
rabphilin 3A |
| chr2_+_121120070 | 9.38 |
ENSMUST00000094639.10
|
Map1a
|
microtubule-associated protein 1 A |
| chr9_+_27702243 | 9.27 |
ENSMUST00000115243.9
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr14_+_74878280 | 9.01 |
ENSMUST00000036653.5
|
Htr2a
|
5-hydroxytryptamine (serotonin) receptor 2A |
| chr2_+_109522781 | 8.94 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr1_+_152830720 | 8.88 |
ENSMUST00000043313.15
ENSMUST00000186621.2 |
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr14_+_66581818 | 8.79 |
ENSMUST00000118426.8
ENSMUST00000121955.8 ENSMUST00000120229.8 ENSMUST00000134440.2 |
Stmn4
|
stathmin-like 4 |
| chr13_-_110417421 | 8.61 |
ENSMUST00000223922.2
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr15_+_82140224 | 8.57 |
ENSMUST00000143238.2
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chrX_+_10351360 | 8.51 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr3_-_88669551 | 8.25 |
ENSMUST00000183267.2
|
Syt11
|
synaptotagmin XI |
| chr6_-_55658242 | 8.19 |
ENSMUST00000044767.10
|
Neurod6
|
neurogenic differentiation 6 |
| chr17_-_46798566 | 8.17 |
ENSMUST00000047034.9
|
Ttbk1
|
tau tubulin kinase 1 |
| chr9_-_29874401 | 7.76 |
ENSMUST00000075069.11
|
Ntm
|
neurotrimin |
| chr15_-_8740218 | 7.75 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr1_-_132635042 | 7.71 |
ENSMUST00000043189.14
|
Nfasc
|
neurofascin |
| chr14_+_70314727 | 7.69 |
ENSMUST00000225200.2
|
Egr3
|
early growth response 3 |
| chr14_-_9015639 | 7.36 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chr9_-_83688294 | 7.33 |
ENSMUST00000034796.14
ENSMUST00000183614.2 |
Elovl4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 |
| chr1_-_132635078 | 7.26 |
ENSMUST00000187861.7
|
Nfasc
|
neurofascin |
| chr3_+_107009896 | 7.19 |
ENSMUST00000196403.2
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr1_+_74582044 | 6.84 |
ENSMUST00000113749.8
ENSMUST00000067916.13 ENSMUST00000113747.8 ENSMUST00000113750.8 |
Plcd4
|
phospholipase C, delta 4 |
| chr12_+_87073338 | 6.81 |
ENSMUST00000110187.8
ENSMUST00000156162.8 |
Tmem63c
|
transmembrane protein 63c |
| chr13_+_83672965 | 6.80 |
ENSMUST00000199432.5
ENSMUST00000198069.5 ENSMUST00000197681.5 ENSMUST00000197722.5 ENSMUST00000197938.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chrX_+_158242121 | 6.80 |
ENSMUST00000112470.3
ENSMUST00000043151.12 ENSMUST00000156172.3 |
Map7d2
|
MAP7 domain containing 2 |
| chr15_-_44978223 | 6.77 |
ENSMUST00000022967.7
|
Kcnv1
|
potassium channel, subfamily V, member 1 |
| chr17_-_90395771 | 6.73 |
ENSMUST00000197268.5
ENSMUST00000173917.8 |
Nrxn1
|
neurexin I |
| chr7_+_91321500 | 6.72 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr5_+_34153328 | 6.68 |
ENSMUST00000056355.9
|
Nat8l
|
N-acetyltransferase 8-like |
| chrX_+_142301572 | 6.66 |
ENSMUST00000033640.14
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr4_+_42158092 | 6.64 |
ENSMUST00000098122.3
|
Gm13306
|
predicted gene 13306 |
| chr14_+_66581745 | 6.63 |
ENSMUST00000152093.8
ENSMUST00000074523.13 |
Stmn4
|
stathmin-like 4 |
| chr15_-_8739893 | 6.61 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr3_-_33137209 | 6.60 |
ENSMUST00000194016.6
ENSMUST00000193681.6 ENSMUST00000192093.6 ENSMUST00000193289.6 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr15_+_74388044 | 6.60 |
ENSMUST00000042035.16
|
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chrX_+_135723531 | 6.58 |
ENSMUST00000113085.2
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr2_+_158452651 | 6.53 |
ENSMUST00000045738.5
|
Slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr2_+_96148418 | 6.52 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr5_-_142594549 | 6.51 |
ENSMUST00000037048.9
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr12_+_87073060 | 6.48 |
ENSMUST00000146292.8
|
Tmem63c
|
transmembrane protein 63c |
| chr9_-_75591274 | 6.48 |
ENSMUST00000214244.2
ENSMUST00000213324.2 ENSMUST00000034699.8 |
Scg3
|
secretogranin III |
| chr17_+_46608333 | 6.44 |
ENSMUST00000188223.7
ENSMUST00000061722.13 ENSMUST00000166280.8 |
Dlk2
|
delta like non-canonical Notch ligand 2 |
| chr10_+_126914755 | 6.43 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr16_+_13804461 | 6.39 |
ENSMUST00000056521.12
ENSMUST00000118412.8 ENSMUST00000131608.2 |
Bmerb1
|
bMERB domain containing 1 |
| chr16_+_6166982 | 6.39 |
ENSMUST00000056416.9
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr16_-_88087087 | 6.28 |
ENSMUST00000211444.2
ENSMUST00000023652.16 ENSMUST00000072256.13 |
Grik1
|
glutamate receptor, ionotropic, kainate 1 |
| chr4_-_41774097 | 6.28 |
ENSMUST00000108036.8
ENSMUST00000108037.9 ENSMUST00000108032.3 ENSMUST00000173865.9 ENSMUST00000155240.2 |
Ccl27a
|
chemokine (C-C motif) ligand 27A |
| chr1_-_79838897 | 6.28 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chrX_+_135723420 | 6.27 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr4_+_9269285 | 6.24 |
ENSMUST00000038841.14
|
Clvs1
|
clavesin 1 |
| chr11_-_69496655 | 6.19 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr14_-_70864448 | 6.18 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr16_+_41353360 | 6.17 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr15_+_38740784 | 6.13 |
ENSMUST00000226440.3
ENSMUST00000239553.1 |
Baalc
|
brain and acute leukemia, cytoplasmic |
| chr3_+_107008343 | 6.13 |
ENSMUST00000197470.5
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr8_+_70536103 | 6.12 |
ENSMUST00000007738.11
|
Hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr6_-_28831746 | 6.06 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr3_+_94385602 | 6.05 |
ENSMUST00000199884.5
ENSMUST00000198316.5 ENSMUST00000197558.5 |
Celf3
|
CUGBP, Elav-like family member 3 |
| chr15_+_97990431 | 6.02 |
ENSMUST00000229280.2
ENSMUST00000163507.8 ENSMUST00000230445.2 |
Pfkm
|
phosphofructokinase, muscle |
| chr2_+_102489558 | 6.01 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_-_52448552 | 6.00 |
ENSMUST00000102760.10
ENSMUST00000102761.9 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr2_-_57942844 | 5.98 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein |
| chr3_-_127202693 | 5.91 |
ENSMUST00000182078.9
|
Ank2
|
ankyrin 2, brain |
| chr6_-_115228800 | 5.89 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr3_-_26187883 | 5.88 |
ENSMUST00000108308.10
ENSMUST00000075054.10 |
Nlgn1
|
neuroligin 1 |
| chr8_-_94262335 | 5.83 |
ENSMUST00000212009.2
ENSMUST00000077816.7 |
Ces5a
|
carboxylesterase 5A |
| chr11_-_119438569 | 5.80 |
ENSMUST00000026670.5
|
Nptx1
|
neuronal pentraxin 1 |
| chr14_+_55173696 | 5.79 |
ENSMUST00000037814.8
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr11_-_97913420 | 5.77 |
ENSMUST00000103144.10
ENSMUST00000017552.13 ENSMUST00000092736.11 ENSMUST00000107562.2 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr4_+_123077515 | 5.76 |
ENSMUST00000152194.2
|
Hpcal4
|
hippocalcin-like 4 |
| chr8_+_66838927 | 5.75 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr9_+_27210500 | 5.74 |
ENSMUST00000214357.2
ENSMUST00000115247.8 ENSMUST00000133213.3 |
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr1_-_25868592 | 5.74 |
ENSMUST00000135518.8
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr14_-_55354392 | 5.70 |
ENSMUST00000022819.13
|
Jph4
|
junctophilin 4 |
| chr7_-_74958121 | 5.66 |
ENSMUST00000085164.7
|
Sv2b
|
synaptic vesicle glycoprotein 2 b |
| chr9_+_74978429 | 5.64 |
ENSMUST00000123128.8
|
Myo5a
|
myosin VA |
| chr11_-_102787950 | 5.64 |
ENSMUST00000067444.10
|
Gfap
|
glial fibrillary acidic protein |
| chr14_-_20027279 | 5.63 |
ENSMUST00000160013.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr4_-_126647156 | 5.60 |
ENSMUST00000030637.14
ENSMUST00000106116.2 |
Ncdn
|
neurochondrin |
| chr1_-_52271455 | 5.59 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr3_-_33137165 | 5.58 |
ENSMUST00000078226.10
ENSMUST00000108224.8 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr13_+_19132375 | 5.55 |
ENSMUST00000239207.2
ENSMUST00000003345.10 ENSMUST00000200466.5 |
Amph
|
amphiphysin |
| chr2_-_25471703 | 5.54 |
ENSMUST00000114217.3
ENSMUST00000191602.2 |
Ajm1
|
apical junction component 1 |
| chr11_+_69016722 | 5.54 |
ENSMUST00000021268.9
|
Aloxe3
|
arachidonate lipoxygenase 3 |
| chr17_+_35454049 | 5.51 |
ENSMUST00000130992.2
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr7_+_126550009 | 5.45 |
ENSMUST00000106332.3
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr2_+_61876956 | 5.44 |
ENSMUST00000112480.3
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr3_-_108062172 | 5.43 |
ENSMUST00000062028.8
|
Gpr61
|
G protein-coupled receptor 61 |
| chr3_-_107366868 | 5.39 |
ENSMUST00000009617.10
ENSMUST00000238670.2 |
Kcnc4
|
potassium voltage gated channel, Shaw-related subfamily, member 4 |
| chr10_-_102326286 | 5.38 |
ENSMUST00000020040.5
|
Nts
|
neurotensin |
| chr16_+_17093941 | 5.36 |
ENSMUST00000164950.11
|
Tmem191c
|
transmembrane protein 191C |
| chr6_-_126621751 | 5.35 |
ENSMUST00000055168.5
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 |
| chr2_+_61876923 | 5.35 |
ENSMUST00000054484.15
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr6_+_124973644 | 5.28 |
ENSMUST00000032479.11
|
Pianp
|
PILR alpha associated neural protein |
| chr6_+_41523664 | 5.28 |
ENSMUST00000103299.3
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr2_+_61876806 | 5.28 |
ENSMUST00000102735.10
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr14_+_55173936 | 5.27 |
ENSMUST00000227441.2
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr7_+_87233554 | 5.26 |
ENSMUST00000125009.9
|
Grm5
|
glutamate receptor, metabotropic 5 |
| chr9_-_112016966 | 5.25 |
ENSMUST00000178410.2
ENSMUST00000172380.10 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr14_-_20027219 | 5.21 |
ENSMUST00000055100.14
ENSMUST00000162425.8 |
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr7_+_126549692 | 5.20 |
ENSMUST00000106335.8
ENSMUST00000146017.3 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr5_+_137059127 | 5.17 |
ENSMUST00000041543.9
ENSMUST00000186451.2 |
Vgf
|
VGF nerve growth factor inducible |
| chr2_-_32737238 | 5.17 |
ENSMUST00000050000.16
|
Stxbp1
|
syntaxin binding protein 1 |
| chr12_-_4891435 | 5.16 |
ENSMUST00000219880.2
ENSMUST00000020964.7 |
Fkbp1b
|
FK506 binding protein 1b |
| chr12_+_76884182 | 5.14 |
ENSMUST00000041008.10
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr2_+_65499097 | 5.13 |
ENSMUST00000200829.4
|
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr2_-_32737208 | 5.13 |
ENSMUST00000077458.7
ENSMUST00000208840.2 |
Stxbp1
|
syntaxin binding protein 1 |
| chr19_-_5148506 | 5.11 |
ENSMUST00000025805.8
|
Cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr10_-_79973210 | 5.11 |
ENSMUST00000170219.9
ENSMUST00000169546.9 |
Cbarp
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein |
| chr9_+_107828136 | 5.06 |
ENSMUST00000049348.9
ENSMUST00000194271.2 |
Traip
|
TRAF-interacting protein |
| chr18_-_43820759 | 5.06 |
ENSMUST00000082254.8
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr12_+_88689638 | 5.04 |
ENSMUST00000190626.7
ENSMUST00000167103.8 |
Nrxn3
|
neurexin III |
| chr7_-_45019984 | 5.04 |
ENSMUST00000003971.10
|
Lin7b
|
lin-7 homolog B (C. elegans) |
| chr18_+_36098090 | 5.03 |
ENSMUST00000176873.8
ENSMUST00000177432.8 ENSMUST00000175734.2 |
Psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr3_+_31204069 | 5.01 |
ENSMUST00000046174.8
|
Cldn11
|
claudin 11 |
| chr17_-_91400142 | 5.00 |
ENSMUST00000160800.9
|
Nrxn1
|
neurexin I |
| chr2_+_91757594 | 4.99 |
ENSMUST00000045537.4
|
Chrm4
|
cholinergic receptor, muscarinic 4 |
| chr2_-_6726417 | 4.96 |
ENSMUST00000142941.8
ENSMUST00000150624.9 ENSMUST00000100429.11 ENSMUST00000182879.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr14_-_55150547 | 4.95 |
ENSMUST00000228495.3
ENSMUST00000228119.3 ENSMUST00000050772.10 ENSMUST00000231305.2 |
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17 |
| chr3_-_127202635 | 4.95 |
ENSMUST00000182959.8
|
Ank2
|
ankyrin 2, brain |
| chr6_-_126621770 | 4.92 |
ENSMUST00000203094.2
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 |
| chr7_+_54485336 | 4.91 |
ENSMUST00000082373.8
|
Luzp2
|
leucine zipper protein 2 |
| chr7_-_28868072 | 4.88 |
ENSMUST00000048923.7
|
Spred3
|
sprouty-related EVH1 domain containing 3 |
| chr7_-_105217851 | 4.87 |
ENSMUST00000188368.7
ENSMUST00000187057.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr9_-_95288775 | 4.87 |
ENSMUST00000036267.8
|
Chst2
|
carbohydrate sulfotransferase 2 |
| chr16_+_20514925 | 4.84 |
ENSMUST00000128273.2
|
Fam131a
|
family with sequence similarity 131, member A |
| chr1_+_158189831 | 4.83 |
ENSMUST00000193042.6
ENSMUST00000046110.16 |
Astn1
|
astrotactin 1 |
| chr14_-_9015757 | 4.82 |
ENSMUST00000153954.8
|
Synpr
|
synaptoporin |
| chr10_+_39296005 | 4.82 |
ENSMUST00000157009.8
|
Fyn
|
Fyn proto-oncogene |
| chr17_+_26036893 | 4.81 |
ENSMUST00000235694.2
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr3_+_68479578 | 4.81 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr7_+_4693759 | 4.81 |
ENSMUST00000048248.9
|
Brsk1
|
BR serine/threonine kinase 1 |
| chrX_+_142447361 | 4.80 |
ENSMUST00000126592.8
ENSMUST00000156449.8 ENSMUST00000155215.8 ENSMUST00000112865.8 |
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr6_-_124441731 | 4.80 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chr15_-_78090591 | 4.79 |
ENSMUST00000120592.2
|
Pvalb
|
parvalbumin |
| chr10_-_70428611 | 4.77 |
ENSMUST00000162251.8
|
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr8_-_122634418 | 4.75 |
ENSMUST00000045557.10
|
Slc7a5
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
| chrX_+_142301666 | 4.73 |
ENSMUST00000134402.8
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr7_+_126549859 | 4.72 |
ENSMUST00000106333.8
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr9_+_108708939 | 4.72 |
ENSMUST00000192235.2
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr1_+_134121170 | 4.70 |
ENSMUST00000038445.13
ENSMUST00000191577.2 |
Mybph
|
myosin binding protein H |
| chr17_-_90395568 | 4.69 |
ENSMUST00000173222.2
|
Nrxn1
|
neurexin I |
| chr3_-_59127571 | 4.68 |
ENSMUST00000199675.2
ENSMUST00000170388.6 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr15_+_16778187 | 4.68 |
ENSMUST00000026432.8
|
Cdh9
|
cadherin 9 |
| chr19_-_46315543 | 4.67 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr9_-_29323032 | 4.66 |
ENSMUST00000115236.2
|
Ntm
|
neurotrimin |
| chr1_+_66426127 | 4.65 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr1_-_75195127 | 4.65 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr16_-_20972750 | 4.63 |
ENSMUST00000170665.3
|
Teddm3
|
transmembrane epididymal family member 3 |
| chr2_+_148237258 | 4.63 |
ENSMUST00000109962.4
|
Sstr4
|
somatostatin receptor 4 |
| chr2_-_65397850 | 4.63 |
ENSMUST00000238483.2
ENSMUST00000100069.9 |
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr8_-_84184978 | 4.60 |
ENSMUST00000081506.11
|
Scoc
|
short coiled-coil protein |
| chr1_-_79836344 | 4.58 |
ENSMUST00000027467.11
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr11_-_102787972 | 4.53 |
ENSMUST00000077902.5
|
Gfap
|
glial fibrillary acidic protein |
| chr9_-_106343137 | 4.53 |
ENSMUST00000164834.3
|
Gpr62
|
G protein-coupled receptor 62 |
| chr1_-_25868788 | 4.52 |
ENSMUST00000151309.8
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr11_+_97732108 | 4.52 |
ENSMUST00000155954.3
ENSMUST00000164364.2 ENSMUST00000170806.2 |
B230217C12Rik
|
RIKEN cDNA B230217C12 gene |
| chr5_-_129907878 | 4.51 |
ENSMUST00000026617.13
|
Phkg1
|
phosphorylase kinase gamma 1 |
| chr2_+_65451100 | 4.51 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr2_-_32977182 | 4.48 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr17_-_25155868 | 4.48 |
ENSMUST00000115228.9
ENSMUST00000117509.8 ENSMUST00000121723.8 ENSMUST00000119115.8 ENSMUST00000121787.8 ENSMUST00000088345.12 ENSMUST00000120035.8 ENSMUST00000115229.10 ENSMUST00000178969.8 |
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr1_+_153541412 | 4.46 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr4_-_114991174 | 4.44 |
ENSMUST00000051400.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr3_+_94385661 | 4.40 |
ENSMUST00000200342.5
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr3_-_95139472 | 4.39 |
ENSMUST00000196025.5
ENSMUST00000198948.5 |
Mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr6_+_103674695 | 4.38 |
ENSMUST00000205098.2
|
Chl1
|
cell adhesion molecule L1-like |
| chrX_+_100342749 | 4.37 |
ENSMUST00000118111.8
ENSMUST00000130555.8 ENSMUST00000151528.8 |
Nlgn3
|
neuroligin 3 |
| chr6_-_113478779 | 4.37 |
ENSMUST00000101059.4
ENSMUST00000204268.3 ENSMUST00000205170.2 ENSMUST00000205075.2 ENSMUST00000204134.3 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr7_+_91321694 | 4.36 |
ENSMUST00000238608.2
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chrX_-_8072714 | 4.35 |
ENSMUST00000089403.10
ENSMUST00000077595.12 ENSMUST00000089402.10 ENSMUST00000082320.12 |
Porcn
|
porcupine O-acyltransferase |
| chr2_-_79959178 | 4.33 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr1_-_52230062 | 4.31 |
ENSMUST00000156887.8
ENSMUST00000129107.2 |
Gls
|
glutaminase |
| chr2_+_132623198 | 4.31 |
ENSMUST00000028826.4
|
Chgb
|
chromogranin B |
| chr15_+_92059224 | 4.30 |
ENSMUST00000068378.6
|
Cntn1
|
contactin 1 |
| chr18_-_25302064 | 4.29 |
ENSMUST00000115817.3
|
Tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr7_-_105218472 | 4.27 |
ENSMUST00000187683.7
ENSMUST00000210079.2 ENSMUST00000187051.7 ENSMUST00000189265.7 ENSMUST00000190369.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr2_+_14878480 | 4.27 |
ENSMUST00000114719.7
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr8_+_73373304 | 4.25 |
ENSMUST00000093427.11
|
Nwd1
|
NACHT and WD repeat domain containing 1 |
| chr15_-_79718423 | 4.25 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chrX_+_133195974 | 4.24 |
ENSMUST00000037687.8
|
Tmem35a
|
transmembrane protein 35A |
| chr16_-_4340920 | 4.23 |
ENSMUST00000090500.10
ENSMUST00000023161.8 |
Srl
|
sarcalumenin |
| chr3_-_33039225 | 4.23 |
ENSMUST00000108221.2
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr3_-_50398027 | 4.22 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr16_+_29028860 | 4.22 |
ENSMUST00000162747.8
|
Plaat1
|
phospholipase A and acyltransferase 1 |
| chr10_-_33500583 | 4.21 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr16_-_94798509 | 4.16 |
ENSMUST00000095873.12
ENSMUST00000099508.4 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr6_-_82916655 | 4.16 |
ENSMUST00000000641.15
|
Sema4f
|
sema domain, immunoglobulin domain (Ig), TM domain, and short cytoplasmic domain |
| chr8_+_24159669 | 4.14 |
ENSMUST00000042352.11
ENSMUST00000207301.2 |
Zmat4
|
zinc finger, matrin type 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 3.2 | 19.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 3.0 | 24.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 2.9 | 8.7 | GO:0098942 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 2.7 | 16.4 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 2.7 | 16.3 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 2.5 | 12.5 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 2.4 | 7.3 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 2.4 | 19.0 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 2.3 | 11.7 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 2.2 | 11.0 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 2.1 | 8.6 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 2.1 | 10.3 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 2.0 | 17.7 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.8 | 5.4 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 1.8 | 5.3 | GO:1902938 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 1.7 | 6.7 | GO:0051944 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 1.7 | 5.0 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.6 | 11.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 1.5 | 4.6 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 1.5 | 9.1 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 1.5 | 12.1 | GO:0061502 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 1.5 | 15.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.5 | 8.9 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 1.5 | 7.4 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 1.5 | 5.8 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.5 | 14.5 | GO:1902473 | regulation of protein localization to synapse(GO:1902473) |
| 1.4 | 4.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 1.3 | 2.7 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 1.3 | 8.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.3 | 13.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.3 | 3.9 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 1.3 | 13.0 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 1.3 | 6.3 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 1.2 | 6.2 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 1.2 | 9.9 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 1.2 | 9.8 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 1.2 | 4.8 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 1.2 | 6.0 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 1.2 | 6.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.2 | 5.9 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 1.1 | 5.6 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.1 | 13.1 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) |
| 1.1 | 4.2 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 1.1 | 4.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 1.1 | 17.9 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 1.0 | 6.2 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.0 | 4.1 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 1.0 | 6.0 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 1.0 | 4.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.0 | 5.0 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.0 | 12.8 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 1.0 | 28.0 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 1.0 | 3.8 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.9 | 11.1 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.9 | 0.9 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.9 | 8.0 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.9 | 2.6 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.9 | 2.6 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.9 | 4.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.8 | 2.5 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.8 | 5.8 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.8 | 2.5 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.8 | 1.7 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.8 | 3.3 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.8 | 8.8 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.8 | 4.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.8 | 0.8 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.8 | 4.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.8 | 2.3 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.8 | 23.9 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.8 | 6.9 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.8 | 3.8 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.8 | 2.3 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.7 | 7.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.7 | 0.7 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.7 | 2.9 | GO:1904799 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.7 | 5.0 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.7 | 3.6 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.7 | 2.8 | GO:0021750 | cerebellar molecular layer development(GO:0021679) vestibular nucleus development(GO:0021750) |
| 0.7 | 2.7 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.7 | 2.0 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) cloacal septation(GO:0060197) |
| 0.7 | 2.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.7 | 4.6 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.7 | 2.6 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.6 | 5.8 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.6 | 1.9 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.6 | 3.9 | GO:0033602 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of dopamine secretion(GO:0033602) |
| 0.6 | 5.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.6 | 5.6 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.6 | 4.4 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.6 | 6.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.6 | 2.5 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.6 | 3.0 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.6 | 6.0 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.6 | 1.7 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.6 | 5.7 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.6 | 1.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.6 | 6.8 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.6 | 2.3 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.6 | 2.2 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.6 | 3.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.6 | 0.6 | GO:0002458 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) |
| 0.5 | 1.6 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.5 | 20.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.5 | 3.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.5 | 1.6 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.5 | 1.6 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.5 | 20.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.5 | 4.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.5 | 3.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.5 | 2.0 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.5 | 2.5 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.5 | 1.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.5 | 18.1 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.5 | 8.2 | GO:0043084 | penile erection(GO:0043084) |
| 0.5 | 2.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.5 | 37.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.5 | 2.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.5 | 1.4 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.5 | 0.9 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.5 | 6.4 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.5 | 1.8 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.5 | 2.3 | GO:0015812 | gamma-aminobutyric acid secretion(GO:0014051) gamma-aminobutyric acid transport(GO:0015812) |
| 0.5 | 1.4 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.5 | 1.4 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.4 | 1.3 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.4 | 2.2 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.4 | 2.7 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.4 | 2.7 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.4 | 1.3 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.4 | 2.6 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.4 | 15.2 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.4 | 6.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.4 | 2.9 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.4 | 1.6 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.4 | 2.8 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.4 | 1.2 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 1.2 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.4 | 1.6 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.4 | 1.9 | GO:0035989 | tendon development(GO:0035989) |
| 0.4 | 1.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.4 | 13.5 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.4 | 4.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.4 | 1.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.4 | 1.5 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.4 | 2.2 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.4 | 2.9 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.4 | 1.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.4 | 1.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.4 | 4.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.4 | 1.4 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.4 | 1.8 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.3 | 1.7 | GO:0097104 | postsynaptic membrane assembly(GO:0097104) |
| 0.3 | 3.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 3.8 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.3 | 1.7 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.3 | 5.1 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.3 | 2.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.3 | 1.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.3 | 2.4 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.3 | 5.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 2.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.3 | 4.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 8.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.3 | 1.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 21.0 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.3 | 5.8 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.3 | 2.5 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.3 | 3.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.3 | 1.6 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.3 | 1.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.3 | 0.9 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 | 4.6 | GO:0030800 | negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cAMP metabolic process(GO:0030815) |
| 0.3 | 3.5 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.3 | 11.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 4.6 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.3 | 2.0 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.3 | 5.7 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.3 | 8.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.3 | 1.1 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.3 | 1.1 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.3 | 1.1 | GO:0070268 | cornification(GO:0070268) |
| 0.3 | 1.1 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.3 | 2.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 1.1 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.3 | 1.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.3 | 1.7 | GO:0051661 | cortical microtubule organization(GO:0043622) maintenance of centrosome location(GO:0051661) |
| 0.3 | 1.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 2.8 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 | 13.5 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.3 | 6.3 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.3 | 2.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.3 | 0.8 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.3 | 3.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.3 | 1.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.3 | 2.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 1.8 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.3 | 0.5 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.3 | 3.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.3 | 1.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.3 | 2.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 0.5 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.3 | 1.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 5.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.2 | 6.9 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.2 | 0.7 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.2 | 3.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 1.9 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 0.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.2 | 6.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.2 | 0.9 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.2 | 1.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.2 | 3.4 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.2 | 0.7 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 3.1 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.2 | 2.9 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 1.3 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.2 | 1.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.2 | 0.9 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.2 | 2.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.2 | 3.8 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.2 | 2.3 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.2 | 1.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 1.5 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.2 | 1.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 1.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.2 | 4.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 3.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.2 | 9.9 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.2 | 0.6 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 | 4.0 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 3.0 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 2.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 7.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.2 | 5.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 | 1.9 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 1.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 0.8 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 | 1.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 3.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.2 | 2.4 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.2 | 4.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 1.6 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.2 | 4.2 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.2 | 9.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 16.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 1.1 | GO:1990166 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.2 | 0.9 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 0.5 | GO:0044861 | protein localization to plasma membrane raft(GO:0044860) protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 1.6 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 6.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 2.2 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 0.7 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.2 | 1.9 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.2 | 0.5 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 2.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 1.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 2.2 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.2 | 0.8 | GO:0018214 | protein carboxylation(GO:0018214) |
| 0.2 | 3.5 | GO:0007379 | segment specification(GO:0007379) |
| 0.2 | 2.5 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.2 | 0.8 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 10.5 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.2 | 1.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 1.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 1.7 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 1.4 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.2 | 0.6 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 6.7 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 2.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 3.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 16.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 4.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 0.4 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 1.0 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 4.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.7 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 4.2 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 3.7 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 0.6 | GO:1902308 | regulation of peptidyl-serine dephosphorylation(GO:1902308) |
| 0.1 | 2.2 | GO:0021895 | cerebral cortex neuron differentiation(GO:0021895) |
| 0.1 | 6.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 2.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 1.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.7 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 1.6 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 7.3 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.1 | 0.8 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.9 | GO:0097324 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.5 | GO:1904046 | protein localization to nuclear pore(GO:0090204) negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.1 | 0.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.6 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 3.2 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.3 | GO:0035700 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.1 | 1.1 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.9 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 2.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 4.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.7 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 1.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 4.3 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 9.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 2.0 | GO:0061615 | glycolytic process through fructose-6-phosphate(GO:0061615) |
| 0.1 | 3.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.2 | GO:0060318 | regulation of definitive erythrocyte differentiation(GO:0010724) definitive erythrocyte differentiation(GO:0060318) |
| 0.1 | 1.8 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.5 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 2.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 4.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.8 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.7 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.7 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 6.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 3.3 | GO:0060078 | regulation of postsynaptic membrane potential(GO:0060078) |
| 0.1 | 6.8 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.1 | 4.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 0.8 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 | 1.6 | GO:0051900 | regulation of mitochondrial depolarization(GO:0051900) |
| 0.1 | 14.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.6 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 1.0 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.1 | 0.9 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.8 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 3.5 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 0.5 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 2.1 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.1 | 0.4 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 | 1.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 3.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 2.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.4 | GO:2000551 | regulation of T-helper 2 cell cytokine production(GO:2000551) |
| 0.1 | 1.0 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 0.2 | GO:1901626 | regulation of postsynaptic membrane organization(GO:1901626) |
| 0.1 | 0.4 | GO:0051882 | mitochondrial depolarization(GO:0051882) |
| 0.1 | 1.3 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.2 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.1 | 1.6 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 2.0 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 2.1 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 1.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 1.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 3.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 2.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 1.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.1 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 8.3 | GO:0015992 | proton transport(GO:0015992) |
| 0.1 | 1.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.4 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.4 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 | 0.4 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 4.9 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.1 | 3.1 | GO:0030816 | positive regulation of cAMP metabolic process(GO:0030816) |
| 0.1 | 0.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.5 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.2 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.1 | 1.8 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.1 | 0.2 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 1.0 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.5 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.7 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 1.0 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 2.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 3.0 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 3.3 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.1 | 0.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.6 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 | 1.5 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 1.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.4 | GO:0044828 | negative regulation by host of viral process(GO:0044793) negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.2 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.1 | 1.7 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 1.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.4 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 1.2 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.1 | 0.7 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 1.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.8 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 1.0 | GO:0045061 | thymic T cell selection(GO:0045061) |
| 0.1 | 6.8 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 1.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 1.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 1.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 5.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.9 | GO:0044146 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.1 | 0.3 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.6 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.9 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 1.9 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 2.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.0 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.1 | 4.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 3.6 | GO:0050808 | synapse organization(GO:0050808) |
| 0.1 | 0.4 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 0.3 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.4 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 2.1 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.9 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) female courtship behavior(GO:0008050) |
| 0.0 | 0.9 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 1.7 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 2.8 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 2.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 3.4 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 1.1 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.6 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 1.5 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.9 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.0 | 1.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.9 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.6 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.1 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.8 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 1.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 5.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.3 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.0 | GO:0090298 | regulation of mitochondrial DNA replication(GO:0090296) negative regulation of mitochondrial DNA replication(GO:0090298) regulation of mitochondrial DNA metabolic process(GO:1901858) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.0 | 0.6 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.5 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 1.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 2.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.8 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 2.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 1.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.2 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.3 | GO:1903541 | regulation of exosomal secretion(GO:1903541) positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 1.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.7 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.4 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 2.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.3 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.5 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 8.1 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.2 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.2 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.0 | GO:1901977 | negative regulation of cell cycle checkpoint(GO:1901977) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 1.1 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.3 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.7 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 2.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 1.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 2.0 | GO:0048515 | spermatid differentiation(GO:0048515) |
| 0.0 | 0.1 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.3 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 1.5 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.4 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.3 | GO:0061371 | determination of heart left/right asymmetry(GO:0061371) |
| 0.0 | 0.5 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.2 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0042596 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) fear response(GO:0042596) |
| 0.0 | 0.1 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 15.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 2.6 | 20.5 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 2.3 | 20.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 2.0 | 6.0 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 1.6 | 8.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 1.6 | 12.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 1.5 | 4.4 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 1.4 | 24.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.4 | 5.6 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 1.4 | 4.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 1.3 | 4.0 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 1.3 | 6.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.3 | 3.8 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 1.3 | 10.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 1.2 | 43.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 1.1 | 13.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.1 | 27.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.0 | 4.0 | GO:0014802 | terminal cisterna(GO:0014802) |
| 1.0 | 18.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.9 | 6.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.8 | 22.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.8 | 3.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.8 | 5.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.8 | 9.1 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.8 | 5.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.8 | 2.5 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.8 | 3.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.8 | 19.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.8 | 10.9 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.8 | 3.9 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.8 | 6.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.7 | 6.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.7 | 2.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.7 | 6.7 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.7 | 2.2 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.7 | 11.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.7 | 26.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.7 | 5.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.7 | 10.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.7 | 5.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.6 | 4.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.6 | 9.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.6 | 6.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.6 | 17.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.6 | 1.8 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.6 | 3.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.5 | 24.0 | GO:0031430 | M band(GO:0031430) |
| 0.5 | 10.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.5 | 1.9 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.5 | 3.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.5 | 11.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.4 | 5.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.4 | 1.3 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.4 | 9.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 2.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.4 | 20.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 4.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.4 | 6.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.4 | 8.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.4 | 1.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.4 | 15.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 12.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 4.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.3 | 1.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.3 | 0.6 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.3 | 3.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.3 | 0.6 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.3 | 10.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 16.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.3 | 1.3 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.3 | 1.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 2.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.3 | 1.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.3 | 2.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 1.3 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.3 | 90.1 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.3 | 2.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 13.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 48.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 1.7 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 4.4 | GO:0042599 | lamellar body(GO:0042599) |
| 0.2 | 1.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 2.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 4.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 0.6 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 1.2 | GO:1904949 | ATPase complex(GO:1904949) |
| 0.2 | 0.8 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.2 | 3.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 3.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 0.6 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 2.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 0.6 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.2 | 0.2 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 2.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.2 | 2.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 0.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 1.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 2.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 0.7 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 16.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.2 | 27.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 17.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.2 | 39.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 1.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 1.6 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.2 | 0.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 2.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.6 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 3.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 2.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 2.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.4 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 1.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 5.4 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 8.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 3.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.7 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 9.9 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.1 | 11.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 1.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 3.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 9.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 8.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.7 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 1.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 1.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 1.7 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 0.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 3.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 2.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.6 | GO:0033202 | DNA helicase complex(GO:0033202) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 3.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.8 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 1.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.5 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 6.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 4.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 3.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.9 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 3.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 2.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 1.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.9 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 1.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.8 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 0.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 2.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 3.1 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.1 | 1.5 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 44.8 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 7.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 2.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 6.4 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 2.1 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.3 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 3.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 4.0 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 0.7 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 8.1 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 2.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 1.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 4.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 1.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.8 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.9 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 3.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 20.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 3.2 | 19.0 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 3.0 | 12.0 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 3.0 | 8.9 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 2.1 | 25.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 1.9 | 5.6 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 1.8 | 5.3 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 1.6 | 9.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 1.6 | 8.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 1.6 | 22.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 1.5 | 9.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 1.5 | 9.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.5 | 8.9 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 1.4 | 4.3 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 1.4 | 1.4 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 1.4 | 15.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.4 | 4.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 1.3 | 8.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.3 | 4.0 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 1.3 | 6.5 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 1.3 | 19.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.3 | 3.9 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 1.2 | 6.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 1.2 | 31.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.2 | 6.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.2 | 6.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.1 | 3.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 1.1 | 30.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 1.1 | 4.3 | GO:0030519 | snoRNP binding(GO:0030519) |
| 1.0 | 3.1 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 1.0 | 5.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.9 | 12.9 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.9 | 4.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.9 | 6.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.8 | 5.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.8 | 3.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.8 | 4.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.7 | 2.2 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.7 | 22.9 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.7 | 8.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.7 | 11.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.7 | 4.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.7 | 18.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.7 | 3.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.7 | 16.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.7 | 2.0 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.7 | 2.7 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.7 | 4.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.7 | 2.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.7 | 3.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.6 | 4.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.6 | 3.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.6 | 9.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.6 | 9.4 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.6 | 20.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.6 | 7.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.6 | 1.8 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.6 | 4.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.6 | 9.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 2.4 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.6 | 5.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.6 | 2.3 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.6 | 2.9 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.6 | 1.7 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.6 | 2.3 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.6 | 5.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.6 | 14.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.5 | 2.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.5 | 2.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.5 | 36.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.5 | 4.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.5 | 3.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.5 | 4.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.5 | 10.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.5 | 10.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.5 | 3.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.5 | 1.4 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.5 | 4.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.5 | 9.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.5 | 4.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.5 | 1.8 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.5 | 1.8 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.5 | 22.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.4 | 3.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 4.9 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 1.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.4 | 1.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.4 | 11.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.4 | 18.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.4 | 8.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.4 | 6.4 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.4 | 1.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.4 | 11.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 2.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 5.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 1.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.4 | 1.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.3 | 1.0 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.3 | 2.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 1.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.3 | 3.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 1.6 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.3 | 26.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 1.7 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.3 | 7.6 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.3 | 1.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.3 | 4.2 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.3 | 1.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 4.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 5.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 2.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 2.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 2.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.3 | 2.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 0.8 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.3 | 1.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.3 | 2.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.3 | 0.5 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.3 | 5.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 1.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 6.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 2.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 2.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 3.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.2 | 1.4 | GO:0005223 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 3.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 2.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 0.9 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 0.7 | GO:0002046 | opsin binding(GO:0002046) |
| 0.2 | 8.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 1.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 3.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.2 | 8.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 1.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 3.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 26.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 9.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 2.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 8.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.2 | 2.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 1.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.2 | 0.6 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 18.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 4.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 13.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 2.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 8.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 5.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.2 | 3.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 0.9 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 0.7 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.2 | 0.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 17.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 5.0 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.2 | 0.7 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 7.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 9.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 2.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 2.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.2 | 1.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.2 | 3.6 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.2 | 1.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 2.7 | GO:0004143 | lipid kinase activity(GO:0001727) diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 0.6 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.2 | 1.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.5 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 1.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 0.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 1.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.7 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 1.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 4.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 3.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.3 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 1.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 4.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 5.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 2.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 42.1 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 2.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 8.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.1 | GO:0022821 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.1 | 1.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 2.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 4.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 1.9 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.1 | 1.7 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.1 | 1.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.7 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.4 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.8 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 2.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 3.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 1.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.7 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 1.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.8 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 2.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 1.6 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.1 | 3.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 1.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 8.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.5 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.1 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.5 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 4.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 5.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 3.4 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 4.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 15.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 5.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 5.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 7.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 4.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 4.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 3.8 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 1.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 2.3 | GO:0005261 | cation channel activity(GO:0005261) |
| 0.0 | 2.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 12.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 6.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.9 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 5.4 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.2 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.0 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 4.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 13.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 16.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 12.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 5.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 9.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 2.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 5.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 4.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 4.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 5.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 9.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 7.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 10.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 1.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 4.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 5.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 0.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 5.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 5.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.5 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 1.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 5.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 5.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 3.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 3.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 12.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.1 | 24.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.9 | 57.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.6 | 9.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.6 | 11.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 27.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 9.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 6.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.4 | 8.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 14.7 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.3 | 4.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.3 | 15.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 3.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.3 | 10.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 15.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 11.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 5.9 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.3 | 3.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 11.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 4.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 1.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 45.0 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.2 | 7.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.2 | 7.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 3.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 3.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 6.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 3.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 3.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 3.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 1.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 12.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 2.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 4.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 8.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 1.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 10.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 4.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 6.6 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.1 | 0.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 19.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 3.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 2.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 7.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 6.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 5.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 1.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 3.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 3.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |