Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Gata4
Z-value: 1.41
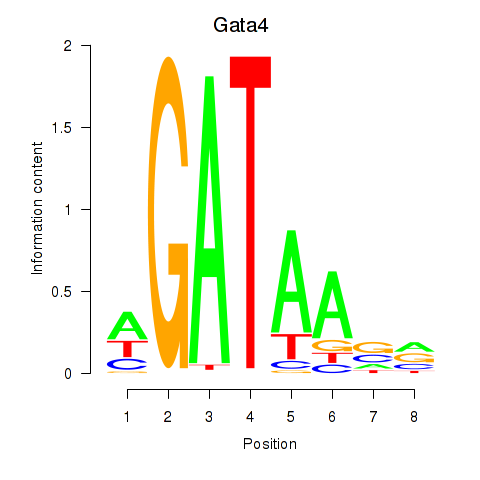
Transcription factors associated with Gata4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata4
|
ENSMUSG00000021944.16 | Gata4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata4 | mm39_v1_chr14_-_63482668_63482714 | 0.61 | 1.7e-08 | Click! |
Activity profile of Gata4 motif
Sorted Z-values of Gata4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_31488208 | 16.03 |
ENSMUST00000001254.6
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr3_+_122688721 | 16.02 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr4_+_119494901 | 13.92 |
ENSMUST00000024015.3
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr19_+_58748132 | 12.60 |
ENSMUST00000026081.5
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr4_+_40920047 | 12.51 |
ENSMUST00000030122.5
|
Spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr2_+_163389068 | 12.51 |
ENSMUST00000109411.8
ENSMUST00000018094.13 |
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr9_+_46139878 | 11.99 |
ENSMUST00000034588.9
ENSMUST00000132155.2 |
Apoa1
|
apolipoprotein A-I |
| chr17_-_84990360 | 11.09 |
ENSMUST00000066175.10
|
Abcg5
|
ATP binding cassette subfamily G member 5 |
| chr9_+_22011488 | 11.02 |
ENSMUST00000213607.2
|
Cnn1
|
calponin 1 |
| chr17_-_26417982 | 10.69 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr10_+_53213763 | 10.16 |
ENSMUST00000219491.2
ENSMUST00000163319.9 ENSMUST00000220197.2 ENSMUST00000046221.8 ENSMUST00000218468.2 ENSMUST00000219921.2 |
Pln
|
phospholamban |
| chr3_+_98129463 | 10.13 |
ENSMUST00000029469.5
|
Reg4
|
regenerating islet-derived family, member 4 |
| chr7_-_4525426 | 9.99 |
ENSMUST00000209148.2
ENSMUST00000098859.10 |
Tnni3
|
troponin I, cardiac 3 |
| chr19_-_11058452 | 9.44 |
ENSMUST00000025636.8
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr12_-_76842263 | 9.24 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr7_-_4525793 | 9.19 |
ENSMUST00000140424.8
|
Tnni3
|
troponin I, cardiac 3 |
| chr17_+_84990541 | 8.52 |
ENSMUST00000045714.15
ENSMUST00000171915.2 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr7_+_28240262 | 8.26 |
ENSMUST00000119180.4
|
Sycn
|
syncollin |
| chr17_+_44445659 | 8.18 |
ENSMUST00000239215.2
|
Clic5
|
chloride intracellular channel 5 |
| chr18_+_11052458 | 8.06 |
ENSMUST00000047762.10
|
Gata6
|
GATA binding protein 6 |
| chr13_+_31809774 | 7.97 |
ENSMUST00000042054.3
|
Foxf2
|
forkhead box F2 |
| chr16_+_38182569 | 7.54 |
ENSMUST00000023494.13
ENSMUST00000114739.2 |
Popdc2
|
popeye domain containing 2 |
| chr11_-_6180127 | 7.27 |
ENSMUST00000004505.3
|
Npc1l1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr11_+_70396070 | 6.92 |
ENSMUST00000019063.3
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr2_+_163348728 | 6.92 |
ENSMUST00000143911.8
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr3_-_14873406 | 6.75 |
ENSMUST00000181860.8
ENSMUST00000144327.3 |
Car1
|
carbonic anhydrase 1 |
| chr6_-_68609426 | 6.75 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chrX_+_10118544 | 6.68 |
ENSMUST00000049910.13
|
Otc
|
ornithine transcarbamylase |
| chr7_+_123061497 | 6.20 |
ENSMUST00000033023.10
|
Aqp8
|
aquaporin 8 |
| chr12_+_8062331 | 6.07 |
ENSMUST00000171239.2
|
Apob
|
apolipoprotein B |
| chr4_+_135455427 | 6.06 |
ENSMUST00000102546.4
|
Il22ra1
|
interleukin 22 receptor, alpha 1 |
| chr7_+_123061535 | 6.02 |
ENSMUST00000098056.6
|
Aqp8
|
aquaporin 8 |
| chr8_+_70525546 | 5.90 |
ENSMUST00000110160.9
ENSMUST00000049197.6 |
Tm6sf2
|
transmembrane 6 superfamily member 2 |
| chr2_-_24985137 | 5.90 |
ENSMUST00000114373.8
|
Noxa1
|
NADPH oxidase activator 1 |
| chrX_+_21581135 | 5.71 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr3_+_89043879 | 5.57 |
ENSMUST00000107482.10
ENSMUST00000127058.2 |
Pklr
|
pyruvate kinase liver and red blood cell |
| chr3_+_89043440 | 5.53 |
ENSMUST00000047111.13
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chr1_+_45350698 | 5.47 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chrX_+_10118600 | 5.47 |
ENSMUST00000115528.3
|
Otc
|
ornithine transcarbamylase |
| chr1_-_136888118 | 5.38 |
ENSMUST00000192357.6
ENSMUST00000027649.14 |
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr7_-_135130374 | 5.25 |
ENSMUST00000053716.8
|
Clrn3
|
clarin 3 |
| chr6_+_24597724 | 5.00 |
ENSMUST00000031694.8
|
Lmod2
|
leiomodin 2 (cardiac) |
| chr6_+_41279199 | 4.91 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr2_-_103133503 | 4.90 |
ENSMUST00000111176.9
|
Ehf
|
ets homologous factor |
| chr14_-_63509131 | 4.88 |
ENSMUST00000132122.2
|
Gata4
|
GATA binding protein 4 |
| chr14_-_55204383 | 4.77 |
ENSMUST00000111456.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr7_+_43284131 | 4.76 |
ENSMUST00000032663.10
|
Ceacam18
|
carcinoembryonic antigen-related cell adhesion molecule 18 |
| chr10_-_116732813 | 4.68 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr14_-_55204023 | 4.64 |
ENSMUST00000124930.8
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr7_-_98887770 | 4.54 |
ENSMUST00000064231.8
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr6_+_30639217 | 4.53 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr6_-_142647944 | 4.43 |
ENSMUST00000100827.5
ENSMUST00000087527.11 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr17_-_35954573 | 4.33 |
ENSMUST00000095467.4
|
Mucl3
|
mucin like 3 |
| chr5_+_115604321 | 4.33 |
ENSMUST00000145785.8
ENSMUST00000031495.11 ENSMUST00000112071.8 ENSMUST00000125568.2 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr19_-_39801188 | 4.32 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr2_-_103133524 | 4.19 |
ENSMUST00000090475.10
|
Ehf
|
ets homologous factor |
| chrX_+_139808351 | 4.13 |
ENSMUST00000033806.5
|
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr14_-_55204092 | 4.07 |
ENSMUST00000081857.14
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr2_+_90948481 | 4.06 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr4_+_150233362 | 4.02 |
ENSMUST00000059893.8
|
Slc2a7
|
solute carrier family 2 (facilitated glucose transporter), member 7 |
| chr14_-_55204054 | 4.00 |
ENSMUST00000226297.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr19_+_27299939 | 3.88 |
ENSMUST00000056708.4
|
Kcnv2
|
potassium channel, subfamily V, member 2 |
| chr2_-_75534985 | 3.83 |
ENSMUST00000102672.5
|
Nfe2l2
|
nuclear factor, erythroid derived 2, like 2 |
| chr1_+_151220222 | 3.83 |
ENSMUST00000023918.13
ENSMUST00000111887.10 ENSMUST00000097543.8 |
Ivns1abp
|
influenza virus NS1A binding protein |
| chr10_+_75983285 | 3.81 |
ENSMUST00000020450.4
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr10_+_69055215 | 3.76 |
ENSMUST00000172261.3
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr1_+_135746330 | 3.75 |
ENSMUST00000038760.10
|
Lad1
|
ladinin |
| chr6_+_68402550 | 3.63 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr14_-_54648057 | 3.60 |
ENSMUST00000200545.2
ENSMUST00000227967.2 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr8_+_4375212 | 3.55 |
ENSMUST00000127460.8
ENSMUST00000136191.8 |
Ccl25
|
chemokine (C-C motif) ligand 25 |
| chr8_+_46081213 | 3.48 |
ENSMUST00000130850.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr17_-_29134288 | 3.47 |
ENSMUST00000062357.6
|
Bnip5
|
BCL2 interacting protein 5 |
| chr19_-_10655391 | 3.44 |
ENSMUST00000025647.7
|
Pga5
|
pepsinogen 5, group I |
| chr14_-_54647647 | 3.23 |
ENSMUST00000228488.2
ENSMUST00000195970.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chrX_+_111221031 | 3.12 |
ENSMUST00000026599.10
ENSMUST00000113415.2 |
Apool
|
apolipoprotein O-like |
| chr17_-_31363245 | 3.08 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr10_+_61484331 | 2.80 |
ENSMUST00000020286.7
|
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr19_+_58658838 | 2.76 |
ENSMUST00000238108.2
|
Pnlip
|
pancreatic lipase |
| chr2_+_15531281 | 2.67 |
ENSMUST00000146205.3
|
Malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr19_+_58658779 | 2.65 |
ENSMUST00000057270.9
|
Pnlip
|
pancreatic lipase |
| chr3_-_107129038 | 2.61 |
ENSMUST00000029504.9
|
Cym
|
chymosin |
| chr5_-_83502966 | 2.54 |
ENSMUST00000053543.11
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr5_+_90920353 | 2.49 |
ENSMUST00000202625.2
|
Pf4
|
platelet factor 4 |
| chr3_+_108272205 | 2.46 |
ENSMUST00000090563.7
|
Mybphl
|
myosin binding protein H-like |
| chrX_+_55825033 | 2.46 |
ENSMUST00000114772.9
ENSMUST00000114768.10 ENSMUST00000155882.8 |
Fhl1
|
four and a half LIM domains 1 |
| chr6_+_87405968 | 2.45 |
ENSMUST00000032125.7
|
Bmp10
|
bone morphogenetic protein 10 |
| chr1_-_43235914 | 2.42 |
ENSMUST00000187357.2
|
Fhl2
|
four and a half LIM domains 2 |
| chr5_-_73349191 | 2.41 |
ENSMUST00000176910.3
|
Fryl
|
FRY like transcription coactivator |
| chr15_-_83316995 | 2.41 |
ENSMUST00000165095.9
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr17_-_15596230 | 2.41 |
ENSMUST00000014917.8
|
Dll1
|
delta like canonical Notch ligand 1 |
| chr7_+_80707328 | 2.39 |
ENSMUST00000107348.2
|
Alpk3
|
alpha-kinase 3 |
| chr6_-_41291634 | 2.38 |
ENSMUST00000064324.12
|
Try5
|
trypsin 5 |
| chr13_+_42454922 | 2.34 |
ENSMUST00000021796.9
|
Edn1
|
endothelin 1 |
| chr15_-_83317020 | 2.30 |
ENSMUST00000231184.2
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr10_-_18887701 | 2.29 |
ENSMUST00000105527.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr5_-_83502793 | 2.28 |
ENSMUST00000146669.2
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr10_-_59452489 | 2.27 |
ENSMUST00000020312.13
|
Mcu
|
mitochondrial calcium uniporter |
| chr17_-_35285146 | 2.21 |
ENSMUST00000174190.2
ENSMUST00000097337.8 |
Mpig6b
|
megakaryocyte and platelet inhibitory receptor G6b |
| chr13_+_41040657 | 2.16 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr6_-_125357756 | 2.05 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr3_-_63758672 | 2.02 |
ENSMUST00000162269.9
ENSMUST00000159676.9 ENSMUST00000175947.8 |
Plch1
|
phospholipase C, eta 1 |
| chrX_-_94521712 | 2.01 |
ENSMUST00000033549.3
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr5_+_64969679 | 1.99 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr6_+_129374260 | 1.99 |
ENSMUST00000032262.14
|
Clec1b
|
C-type lectin domain family 1, member b |
| chr1_-_74924481 | 1.98 |
ENSMUST00000159232.2
ENSMUST00000068631.4 |
Fev
|
FEV transcription factor, ETS family member |
| chr2_-_153083322 | 1.95 |
ENSMUST00000056924.14
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr14_-_70585874 | 1.94 |
ENSMUST00000152067.8
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr19_+_24853039 | 1.93 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chrX_+_55824797 | 1.92 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr15_+_55171138 | 1.92 |
ENSMUST00000023053.12
ENSMUST00000110217.10 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr7_-_98829474 | 1.90 |
ENSMUST00000207611.2
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr4_+_124696394 | 1.89 |
ENSMUST00000175875.2
|
Mtf1
|
metal response element binding transcription factor 1 |
| chr16_+_29884153 | 1.85 |
ENSMUST00000023171.8
|
Hes1
|
hes family bHLH transcription factor 1 |
| chr6_+_125529911 | 1.85 |
ENSMUST00000112254.8
ENSMUST00000112253.6 |
Vwf
|
Von Willebrand factor |
| chr1_+_174000304 | 1.84 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr4_-_137157824 | 1.81 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr16_+_44913974 | 1.76 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr4_+_124696336 | 1.75 |
ENSMUST00000138807.8
ENSMUST00000030723.3 |
Mtf1
|
metal response element binding transcription factor 1 |
| chr14_+_33662976 | 1.72 |
ENSMUST00000100720.2
|
Gdf2
|
growth differentiation factor 2 |
| chr6_+_17306414 | 1.71 |
ENSMUST00000150901.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr13_-_23650045 | 1.70 |
ENSMUST00000041674.14
ENSMUST00000110434.2 |
Btn1a1
|
butyrophilin, subfamily 1, member A1 |
| chr13_-_116446166 | 1.55 |
ENSMUST00000036060.13
|
Isl1
|
ISL1 transcription factor, LIM/homeodomain |
| chr6_+_17306379 | 1.53 |
ENSMUST00000115455.3
|
Cav1
|
caveolin 1, caveolae protein |
| chr6_+_115398996 | 1.48 |
ENSMUST00000000450.5
|
Pparg
|
peroxisome proliferator activated receptor gamma |
| chr10_+_116013256 | 1.46 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr9_+_21176582 | 1.44 |
ENSMUST00000065005.5
|
Atg4d
|
autophagy related 4D, cysteine peptidase |
| chr18_-_44308126 | 1.42 |
ENSMUST00000066328.5
|
Spinkl
|
serine protease inhibitor, Kazal type-like |
| chr6_+_129374441 | 1.40 |
ENSMUST00000112081.9
ENSMUST00000112079.3 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr3_+_101917455 | 1.40 |
ENSMUST00000066187.6
ENSMUST00000198675.2 |
Nhlh2
|
nescient helix loop helix 2 |
| chr3_+_84500854 | 1.32 |
ENSMUST00000062623.4
|
Tigd4
|
tigger transposable element derived 4 |
| chr3_-_88460166 | 1.30 |
ENSMUST00000119002.2
ENSMUST00000029698.15 |
Lamtor2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr19_-_6065415 | 1.29 |
ENSMUST00000237519.2
|
Capn1
|
calpain 1 |
| chr3_+_69914946 | 1.29 |
ENSMUST00000053013.6
|
Otol1
|
otolin 1 |
| chr14_+_55798517 | 1.28 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr17_-_84495364 | 1.25 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr15_+_53748302 | 1.25 |
ENSMUST00000100666.3
|
Gm7489
|
predicted gene 7489 |
| chr6_+_17306334 | 1.24 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr17_-_25394445 | 1.20 |
ENSMUST00000224277.2
|
Percc1
|
proline and glutamate rich with coiled coil 1 |
| chr19_+_55886708 | 1.18 |
ENSMUST00000148666.3
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr7_-_80053063 | 1.17 |
ENSMUST00000147150.2
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr19_-_43512929 | 1.16 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr14_+_55798362 | 1.14 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr9_+_92074989 | 1.10 |
ENSMUST00000179751.3
|
Plscr5
|
phospholipid scramblase family, member 5 |
| chr17_+_37180437 | 1.08 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr8_-_86107593 | 1.04 |
ENSMUST00000122452.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr19_-_6065181 | 1.04 |
ENSMUST00000236537.2
ENSMUST00000025891.11 |
Capn1
|
calpain 1 |
| chr3_+_116653113 | 0.99 |
ENSMUST00000040260.11
|
Frrs1
|
ferric-chelate reductase 1 |
| chr11_+_49404445 | 0.97 |
ENSMUST00000060434.5
|
Olfr1384
|
olfactory receptor 1384 |
| chr13_-_73848807 | 0.95 |
ENSMUST00000022048.6
|
Slc6a19
|
solute carrier family 6 (neurotransmitter transporter), member 19 |
| chr2_+_164245114 | 0.95 |
ENSMUST00000017151.2
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr16_-_92622972 | 0.94 |
ENSMUST00000023673.14
|
Runx1
|
runt related transcription factor 1 |
| chr13_-_56696222 | 0.94 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr5_+_32616187 | 0.93 |
ENSMUST00000015100.15
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr5_+_42225303 | 0.93 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr6_+_30541581 | 0.92 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr1_-_57008986 | 0.91 |
ENSMUST00000176759.2
ENSMUST00000177424.2 |
Satb2
|
special AT-rich sequence binding protein 2 |
| chr4_-_14621669 | 0.90 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr14_+_53220913 | 0.89 |
ENSMUST00000167409.2
|
Trav9d-4
|
T cell receptor alpha variable 9D-4 |
| chr15_-_38518458 | 0.89 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chr4_+_52964547 | 0.88 |
ENSMUST00000215010.2
ENSMUST00000215127.2 |
Olfr270
|
olfactory receptor 270 |
| chr2_+_72306503 | 0.83 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr11_-_87811788 | 0.82 |
ENSMUST00000216461.2
ENSMUST00000217112.2 |
Olfr464
|
olfactory receptor 464 |
| chr5_+_31454939 | 0.80 |
ENSMUST00000201675.3
|
Gckr
|
glucokinase regulatory protein |
| chr10_-_83484576 | 0.79 |
ENSMUST00000020500.14
|
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr15_-_83054369 | 0.77 |
ENSMUST00000162834.3
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr19_-_6065799 | 0.76 |
ENSMUST00000235138.2
|
Capn1
|
calpain 1 |
| chr13_-_56696310 | 0.75 |
ENSMUST00000062806.6
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr3_-_75177378 | 0.74 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr4_-_82623972 | 0.74 |
ENSMUST00000155821.2
|
Nfib
|
nuclear factor I/B |
| chr12_-_28673259 | 0.73 |
ENSMUST00000220836.2
|
Colec11
|
collectin sub-family member 11 |
| chr11_+_82802079 | 0.68 |
ENSMUST00000018989.14
ENSMUST00000164945.3 |
Unc45b
|
unc-45 myosin chaperone B |
| chr15_-_83054698 | 0.67 |
ENSMUST00000162178.8
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr4_-_137137088 | 0.65 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr11_-_46280281 | 0.62 |
ENSMUST00000101306.4
|
Itk
|
IL2 inducible T cell kinase |
| chr3_+_101917392 | 0.61 |
ENSMUST00000196324.2
|
Nhlh2
|
nescient helix loop helix 2 |
| chr6_-_122833109 | 0.58 |
ENSMUST00000042081.9
|
C3ar1
|
complement component 3a receptor 1 |
| chr17_-_35304582 | 0.57 |
ENSMUST00000038507.7
|
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr9_-_44253588 | 0.57 |
ENSMUST00000215091.2
|
Hmbs
|
hydroxymethylbilane synthase |
| chr5_+_104318542 | 0.56 |
ENSMUST00000112771.2
|
Dspp
|
dentin sialophosphoprotein |
| chr10_-_83484467 | 0.55 |
ENSMUST00000146876.9
ENSMUST00000176294.2 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr11_+_101151394 | 0.51 |
ENSMUST00000103108.8
|
Wnk4
|
WNK lysine deficient protein kinase 4 |
| chr6_+_131463363 | 0.50 |
ENSMUST00000159229.3
ENSMUST00000075020.10 |
Gm6619
5430401F13Rik
|
predicted gene 6619 RIKEN cDNA 5430401F13 gene |
| chr8_+_46111361 | 0.49 |
ENSMUST00000210946.2
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr16_+_23338960 | 0.49 |
ENSMUST00000211460.2
ENSMUST00000210658.2 ENSMUST00000209198.2 ENSMUST00000210371.2 ENSMUST00000211499.2 ENSMUST00000210795.2 ENSMUST00000209422.2 |
Gm45338
Rtp4
|
predicted gene 45338 receptor transporter protein 4 |
| chr12_-_91815855 | 0.48 |
ENSMUST00000167466.2
ENSMUST00000021347.12 ENSMUST00000178462.8 |
Sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr18_-_44329234 | 0.46 |
ENSMUST00000239421.2
ENSMUST00000097587.5 |
Spink11
|
serine peptidase inhibitor, Kazal type 11 |
| chr7_-_4633472 | 0.45 |
ENSMUST00000055085.8
|
Tmem86b
|
transmembrane protein 86B |
| chr11_-_99241924 | 0.44 |
ENSMUST00000017732.3
|
Krt27
|
keratin 27 |
| chr7_-_30555592 | 0.43 |
ENSMUST00000185748.2
ENSMUST00000094583.2 |
Ffar3
|
free fatty acid receptor 3 |
| chr19_+_12405268 | 0.43 |
ENSMUST00000168148.2
|
Pfpl
|
pore forming protein-like |
| chrX_-_59449137 | 0.43 |
ENSMUST00000033480.13
ENSMUST00000101527.3 |
Atp11c
|
ATPase, class VI, type 11C |
| chr2_+_85805557 | 0.42 |
ENSMUST00000082191.5
|
Olfr1029
|
olfactory receptor 1029 |
| chr15_-_103160082 | 0.40 |
ENSMUST00000149111.8
ENSMUST00000132836.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr9_-_80347482 | 0.40 |
ENSMUST00000085289.12
ENSMUST00000185068.2 ENSMUST00000113250.10 |
Impg1
|
interphotoreceptor matrix proteoglycan 1 |
| chr4_+_56740070 | 0.40 |
ENSMUST00000181745.2
|
Gm26657
|
predicted gene, 26657 |
| chr17_-_89508103 | 0.39 |
ENSMUST00000035701.6
|
Fshr
|
follicle stimulating hormone receptor |
| chr3_+_79792238 | 0.38 |
ENSMUST00000135021.2
|
Gask1b
|
golgi associated kinase 1B |
| chr10_+_100426346 | 0.38 |
ENSMUST00000218464.2
ENSMUST00000188930.7 |
1700017N19Rik
|
RIKEN cDNA 1700017N19 gene |
| chr18_-_39622295 | 0.37 |
ENSMUST00000131885.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr15_-_103159892 | 0.36 |
ENSMUST00000133600.8
ENSMUST00000134554.2 ENSMUST00000156927.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 19.4 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 4.4 | 17.5 | GO:0007522 | visceral muscle development(GO:0007522) |
| 4.0 | 12.1 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 3.3 | 19.6 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 3.2 | 19.2 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 3.0 | 12.0 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 2.3 | 9.2 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 1.6 | 4.9 | GO:0060464 | lung lobe formation(GO:0060464) diaphragm morphogenesis(GO:0060540) |
| 1.6 | 8.1 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 1.5 | 12.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 1.5 | 4.5 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 1.5 | 7.5 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 1.4 | 4.3 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 1.3 | 3.8 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 1.2 | 11.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 1.1 | 6.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 1.1 | 11.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 1.1 | 5.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.1 | 16.0 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 1.1 | 4.3 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 1.0 | 12.6 | GO:0019374 | galactolipid metabolic process(GO:0019374) |
| 1.0 | 10.2 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 1.0 | 12.7 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.9 | 3.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.8 | 2.4 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.8 | 2.3 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.8 | 2.3 | GO:0002631 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.7 | 4.5 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.7 | 2.7 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.7 | 2.0 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.6 | 1.9 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.6 | 8.6 | GO:0010744 | positive regulation of macrophage derived foam cell differentiation(GO:0010744) |
| 0.5 | 5.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.5 | 1.6 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.5 | 3.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.5 | 3.5 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.5 | 4.7 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.5 | 9.7 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.4 | 11.0 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.4 | 6.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.4 | 1.2 | GO:0006533 | fumarate metabolic process(GO:0006106) glycerol biosynthetic process(GO:0006114) aspartate catabolic process(GO:0006533) |
| 0.4 | 5.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.4 | 1.4 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.3 | 7.5 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.3 | 2.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.3 | 1.2 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.3 | 10.7 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.3 | 2.0 | GO:0051611 | regulation of serotonin uptake(GO:0051611) |
| 0.3 | 5.9 | GO:0060263 | regulation of hydrogen peroxide metabolic process(GO:0010310) regulation of respiratory burst(GO:0060263) |
| 0.3 | 3.8 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.3 | 4.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 1.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 0.9 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.2 | 2.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 3.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 0.9 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 1.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 4.8 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.2 | 0.6 | GO:0071460 | positive regulation of enamel mineralization(GO:0070175) cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 0.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 3.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 2.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 3.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 4.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.4 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.1 | 1.3 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 6.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.2 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.1 | 0.4 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 1.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.3 | GO:0036034 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.1 | 3.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 4.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 0.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.6 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) tolerance induction dependent upon immune response(GO:0002461) |
| 0.1 | 6.1 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 1.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 1.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.1 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.4 | GO:1900170 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 1.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.8 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 2.0 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 1.9 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 1.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 3.8 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.5 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.0 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 1.9 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 9.1 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 1.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 2.4 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.2 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.5 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 7.1 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.5 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 1.5 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 1.2 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 3.5 | GO:0051607 | defense response to virus(GO:0051607) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.2 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 2.7 | 19.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 2.4 | 12.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 2.0 | 6.1 | GO:0034359 | mature chylomicron(GO:0034359) |
| 1.5 | 16.0 | GO:0045179 | apical cortex(GO:0045179) |
| 1.4 | 4.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 1.3 | 19.6 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.8 | 19.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.6 | 5.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.5 | 12.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.5 | 4.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.5 | 16.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.4 | 5.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 5.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 3.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.4 | 5.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 2.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 10.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 2.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 4.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 1.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 2.5 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.2 | 1.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 3.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 8.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.9 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 8.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 2.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.5 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 10.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 2.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 37.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 4.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 9.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 34.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 9.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 19.4 | GO:0070540 | stearic acid binding(GO:0070540) |
| 4.0 | 12.0 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 3.0 | 12.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 2.9 | 17.5 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 2.7 | 19.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 2.3 | 13.9 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 2.2 | 11.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 2.0 | 6.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 1.8 | 16.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.5 | 4.4 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 1.5 | 10.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.1 | 5.7 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 1.1 | 6.4 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 1.0 | 17.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.9 | 10.7 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.9 | 3.5 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.9 | 18.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.8 | 25.7 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.8 | 2.3 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.7 | 4.5 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.7 | 2.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.7 | 12.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.7 | 6.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.6 | 1.8 | GO:0071820 | N-box binding(GO:0071820) |
| 0.6 | 2.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.5 | 2.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.5 | 2.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.5 | 4.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 5.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 1.2 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.4 | 3.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 2.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.3 | 5.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 5.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 6.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.3 | 3.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.3 | 6.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.3 | 2.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.3 | 9.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 7.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 5.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 7.0 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 0.9 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 6.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 9.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 4.7 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 1.9 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 4.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 2.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 3.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 3.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.8 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 14.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 3.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 8.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 5.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 5.4 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.6 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 0.4 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 1.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 3.0 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 11.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 8.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 1.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 42.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 10.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 12.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 13.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 4.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 7.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 10.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 5.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 5.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 2.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 11.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 3.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 31.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.9 | 12.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.9 | 30.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.6 | 36.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 7.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.4 | 6.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.3 | 24.5 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.3 | 6.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 4.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 4.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 4.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 6.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 2.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 2.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 7.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 10.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 3.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 2.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 7.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |