Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Gata5
Z-value: 1.26
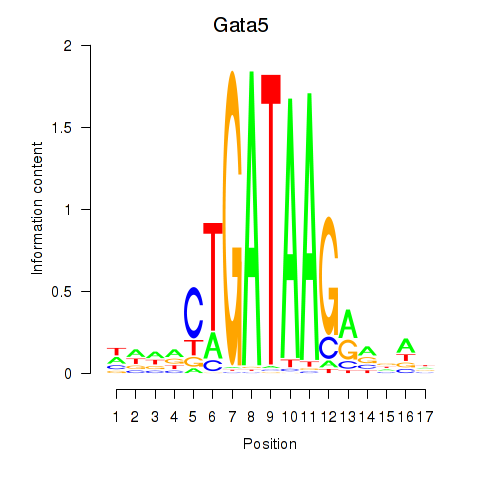
Transcription factors associated with Gata5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata5
|
ENSMUSG00000015627.6 | Gata5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata5 | mm39_v1_chr2_-_179976458_179976509 | 0.33 | 4.2e-03 | Click! |
Activity profile of Gata5 motif
Sorted Z-values of Gata5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_14873406 | 11.63 |
ENSMUST00000181860.8
ENSMUST00000144327.3 |
Car1
|
carbonic anhydrase 1 |
| chr2_+_84810802 | 10.37 |
ENSMUST00000028467.6
|
Prg2
|
proteoglycan 2, bone marrow |
| chr7_-_98887770 | 10.19 |
ENSMUST00000064231.8
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr17_+_41121979 | 9.37 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr6_-_68609426 | 8.91 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr7_-_103477126 | 8.86 |
ENSMUST00000023934.8
|
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr3_-_14843512 | 8.66 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr6_+_68402550 | 8.12 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr17_+_35133435 | 7.98 |
ENSMUST00000007249.15
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr11_+_32233511 | 7.05 |
ENSMUST00000093209.4
|
Hba-a1
|
hemoglobin alpha, adult chain 1 |
| chr11_+_32246489 | 6.84 |
ENSMUST00000093207.4
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr6_+_41498716 | 6.82 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr5_-_87054796 | 6.45 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr4_+_119494901 | 6.30 |
ENSMUST00000024015.3
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr14_+_47605208 | 6.25 |
ENSMUST00000151405.9
|
Lgals3
|
lectin, galactose binding, soluble 3 |
| chr6_+_78347844 | 6.25 |
ENSMUST00000096904.6
ENSMUST00000203266.2 |
Reg3b
|
regenerating islet-derived 3 beta |
| chr7_-_120673761 | 6.19 |
ENSMUST00000047194.4
|
Igsf6
|
immunoglobulin superfamily, member 6 |
| chr17_-_26417982 | 6.16 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr6_+_41279199 | 6.14 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr17_-_34218301 | 5.91 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr15_-_103159892 | 5.73 |
ENSMUST00000133600.8
ENSMUST00000134554.2 ENSMUST00000156927.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr6_+_78347636 | 5.38 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr15_-_78449172 | 5.37 |
ENSMUST00000230952.2
|
Rac2
|
Rac family small GTPase 2 |
| chr6_+_42238891 | 5.33 |
ENSMUST00000095987.4
|
Tmem139
|
transmembrane protein 139 |
| chr16_+_95058417 | 5.26 |
ENSMUST00000113861.8
ENSMUST00000113854.8 ENSMUST00000113862.8 ENSMUST00000037154.14 ENSMUST00000113855.8 |
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr2_+_84818538 | 5.20 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3 |
| chr13_-_100338469 | 5.19 |
ENSMUST00000167986.3
ENSMUST00000117913.8 |
Naip2
|
NLR family, apoptosis inhibitory protein 2 |
| chr16_+_95058505 | 5.15 |
ENSMUST00000113856.8
ENSMUST00000125847.2 ENSMUST00000134166.8 ENSMUST00000140222.8 |
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr3_-_98670369 | 5.13 |
ENSMUST00000107019.8
ENSMUST00000107018.8 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr1_+_40468720 | 5.06 |
ENSMUST00000174335.8
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr15_-_103160082 | 5.00 |
ENSMUST00000149111.8
ENSMUST00000132836.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr18_+_11052458 | 4.98 |
ENSMUST00000047762.10
|
Gata6
|
GATA binding protein 6 |
| chrX_-_8059597 | 4.86 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr13_-_41981893 | 4.63 |
ENSMUST00000137905.2
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr11_-_83177548 | 4.53 |
ENSMUST00000163961.3
|
Slfn14
|
schlafen 14 |
| chr16_+_95058895 | 4.39 |
ENSMUST00000113859.8
ENSMUST00000152516.2 |
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr8_+_105460627 | 4.37 |
ENSMUST00000034346.15
ENSMUST00000164182.3 |
Ces2a
|
carboxylesterase 2A |
| chr8_+_67947494 | 4.16 |
ENSMUST00000093470.7
ENSMUST00000163856.3 |
Nat2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr13_-_41981812 | 4.14 |
ENSMUST00000223337.2
ENSMUST00000221691.2 |
Adtrp
|
androgen dependent TFPI regulating protein |
| chr6_-_69584812 | 4.08 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr11_-_102360664 | 4.08 |
ENSMUST00000103086.4
|
Itga2b
|
integrin alpha 2b |
| chr13_+_108452866 | 4.05 |
ENSMUST00000051594.12
|
Depdc1b
|
DEP domain containing 1B |
| chr15_+_55171138 | 4.02 |
ENSMUST00000023053.12
ENSMUST00000110217.10 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr1_-_131025562 | 3.94 |
ENSMUST00000016672.11
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chrX_+_111404963 | 3.91 |
ENSMUST00000026602.9
ENSMUST00000113412.3 |
2010106E10Rik
|
RIKEN cDNA 2010106E10 gene |
| chr17_+_44445659 | 3.91 |
ENSMUST00000239215.2
|
Clic5
|
chloride intracellular channel 5 |
| chr14_-_51295099 | 3.89 |
ENSMUST00000227764.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr4_-_119047202 | 3.85 |
ENSMUST00000239029.2
ENSMUST00000138395.9 ENSMUST00000156746.3 |
Ermap
|
erythroblast membrane-associated protein |
| chr1_+_127657142 | 3.74 |
ENSMUST00000038006.8
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr6_+_48881913 | 3.71 |
ENSMUST00000162948.7
ENSMUST00000167529.3 |
Aoc1
|
amine oxidase, copper-containing 1 |
| chr13_+_4283729 | 3.48 |
ENSMUST00000081326.7
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr7_-_135130374 | 3.48 |
ENSMUST00000053716.8
|
Clrn3
|
clarin 3 |
| chr3_-_107129038 | 3.43 |
ENSMUST00000029504.9
|
Cym
|
chymosin |
| chr8_-_83128437 | 3.42 |
ENSMUST00000209573.3
|
Il15
|
interleukin 15 |
| chr4_-_137157824 | 3.40 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr1_-_170755136 | 3.39 |
ENSMUST00000046322.14
ENSMUST00000159171.2 |
Fcrla
|
Fc receptor-like A |
| chr4_-_119047167 | 3.36 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chr19_+_11420937 | 3.36 |
ENSMUST00000035258.10
|
Ms4a4b
|
membrane-spanning 4-domains, subfamily A, member 4B |
| chr1_+_51328265 | 3.28 |
ENSMUST00000051572.8
|
Cavin2
|
caveolae associated 2 |
| chr1_+_88015524 | 3.27 |
ENSMUST00000113139.2
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr4_+_135455427 | 3.24 |
ENSMUST00000102546.4
|
Il22ra1
|
interleukin 22 receptor, alpha 1 |
| chr10_+_34359395 | 3.21 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr16_+_58490625 | 3.18 |
ENSMUST00000060077.7
|
Cpox
|
coproporphyrinogen oxidase |
| chrM_+_5319 | 3.17 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr16_+_49675682 | 3.16 |
ENSMUST00000114496.3
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr9_+_46139878 | 3.16 |
ENSMUST00000034588.9
ENSMUST00000132155.2 |
Apoa1
|
apolipoprotein A-I |
| chr2_+_84867554 | 3.09 |
ENSMUST00000077798.13
|
Ssrp1
|
structure specific recognition protein 1 |
| chr6_+_70640233 | 3.00 |
ENSMUST00000103400.3
|
Igkv3-5
|
immunoglobulin kappa chain variable 3-5 |
| chr7_-_103928939 | 2.97 |
ENSMUST00000051795.10
|
Trim5
|
tripartite motif-containing 5 |
| chr8_-_110765983 | 2.94 |
ENSMUST00000109222.4
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr13_-_100454182 | 2.92 |
ENSMUST00000118574.8
|
Naip6
|
NLR family, apoptosis inhibitory protein 6 |
| chr10_-_116732813 | 2.90 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr6_-_68887957 | 2.90 |
ENSMUST00000200454.2
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr7_+_98089623 | 2.85 |
ENSMUST00000206435.2
|
Gucy2d
|
guanylate cyclase 2d |
| chr6_+_38918327 | 2.81 |
ENSMUST00000160963.2
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr10_+_127595639 | 2.79 |
ENSMUST00000128247.2
|
Rdh16f1
|
RDH16 family member 1 |
| chr15_+_80507671 | 2.76 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr1_+_85528392 | 2.76 |
ENSMUST00000080204.11
|
Sp140
|
Sp140 nuclear body protein |
| chr9_+_20940669 | 2.76 |
ENSMUST00000001040.7
ENSMUST00000215077.2 |
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr4_-_119047180 | 2.75 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr11_+_9141934 | 2.74 |
ENSMUST00000042740.13
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr1_-_173703424 | 2.73 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr19_-_32038838 | 2.71 |
ENSMUST00000096119.5
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr4_-_155012643 | 2.61 |
ENSMUST00000123514.8
|
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr3_+_145827410 | 2.61 |
ENSMUST00000039450.5
|
Mcoln3
|
mucolipin 3 |
| chr3_+_142300601 | 2.58 |
ENSMUST00000029936.5
|
Gbp2b
|
guanylate binding protein 2b |
| chr2_+_84867783 | 2.53 |
ENSMUST00000168266.8
ENSMUST00000130729.3 |
Ssrp1
|
structure specific recognition protein 1 |
| chr5_-_35897331 | 2.53 |
ENSMUST00000201511.2
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr4_-_119047146 | 2.49 |
ENSMUST00000124626.9
|
Ermap
|
erythroblast membrane-associated protein |
| chr6_+_41331039 | 2.46 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr6_-_3988835 | 2.44 |
ENSMUST00000203257.2
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr16_+_36755338 | 2.41 |
ENSMUST00000023531.15
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr11_+_96920751 | 2.35 |
ENSMUST00000021249.11
|
Scrn2
|
secernin 2 |
| chr3_-_98669720 | 2.32 |
ENSMUST00000146196.4
|
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr8_-_110766009 | 2.20 |
ENSMUST00000212934.2
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr11_-_114952984 | 2.14 |
ENSMUST00000062787.8
|
Cd300e
|
CD300E molecule |
| chr12_+_102249294 | 2.13 |
ENSMUST00000056950.14
|
Rin3
|
Ras and Rab interactor 3 |
| chr5_-_73349191 | 2.04 |
ENSMUST00000176910.3
|
Fryl
|
FRY like transcription coactivator |
| chr19_-_7779943 | 2.02 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr5_+_137568982 | 2.02 |
ENSMUST00000196471.5
ENSMUST00000198783.5 |
Tfr2
|
transferrin receptor 2 |
| chr9_-_99599312 | 2.00 |
ENSMUST00000112882.9
ENSMUST00000131922.2 |
Cldn18
|
claudin 18 |
| chr8_-_3517617 | 1.96 |
ENSMUST00000111081.10
ENSMUST00000004686.13 |
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr6_+_41512480 | 1.91 |
ENSMUST00000103289.2
ENSMUST00000103290.2 ENSMUST00000193061.2 |
Trbj1-6
Trbj1-7
|
T cell receptor beta joining 1-6 T cell receptor beta joining 1-7 |
| chr11_+_116422570 | 1.90 |
ENSMUST00000106387.9
|
Sphk1
|
sphingosine kinase 1 |
| chr11_+_116422712 | 1.89 |
ENSMUST00000100201.10
|
Sphk1
|
sphingosine kinase 1 |
| chr12_-_114793177 | 1.87 |
ENSMUST00000103511.2
ENSMUST00000195735.2 |
Ighv1-31
|
immunoglobulin heavy variable 1-31 |
| chr1_-_85248264 | 1.86 |
ENSMUST00000093506.12
ENSMUST00000064341.9 |
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr16_-_26810402 | 1.86 |
ENSMUST00000231299.2
|
Gmnc
|
geminin coiled-coil domain containing |
| chr11_+_70529944 | 1.83 |
ENSMUST00000055184.7
ENSMUST00000108551.3 |
Gp1ba
|
glycoprotein 1b, alpha polypeptide |
| chr19_-_7780025 | 1.82 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr6_-_3988900 | 1.81 |
ENSMUST00000183682.3
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr8_-_21392510 | 1.81 |
ENSMUST00000122025.9
|
Gm15056
|
predicted gene 15056 |
| chr11_-_45845992 | 1.75 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr8_+_85428059 | 1.74 |
ENSMUST00000238364.2
ENSMUST00000238562.2 ENSMUST00000037165.6 |
Lyl1
|
lymphoblastomic leukemia 1 |
| chr1_-_155617773 | 1.71 |
ENSMUST00000027740.14
|
Lhx4
|
LIM homeobox protein 4 |
| chr11_+_94827050 | 1.70 |
ENSMUST00000001547.8
|
Col1a1
|
collagen, type I, alpha 1 |
| chr10_-_116786271 | 1.69 |
ENSMUST00000020375.7
ENSMUST00000219109.2 |
Rab3ip
|
RAB3A interacting protein |
| chr11_+_116423266 | 1.67 |
ENSMUST00000106386.8
ENSMUST00000145737.8 ENSMUST00000155102.8 ENSMUST00000063446.13 |
Sphk1
|
sphingosine kinase 1 |
| chr11_+_96920956 | 1.63 |
ENSMUST00000153482.2
|
Scrn2
|
secernin 2 |
| chr7_-_80053063 | 1.53 |
ENSMUST00000147150.2
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr13_+_3887757 | 1.52 |
ENSMUST00000042219.6
|
Calm4
|
calmodulin 4 |
| chr9_+_50405817 | 1.44 |
ENSMUST00000114474.8
ENSMUST00000188047.2 |
Plet1
|
placenta expressed transcript 1 |
| chr15_+_102314578 | 1.41 |
ENSMUST00000170884.8
ENSMUST00000163709.8 |
Sp1
|
trans-acting transcription factor 1 |
| chr7_-_4633472 | 1.40 |
ENSMUST00000055085.8
|
Tmem86b
|
transmembrane protein 86B |
| chr13_+_108452930 | 1.40 |
ENSMUST00000171178.2
|
Depdc1b
|
DEP domain containing 1B |
| chr11_-_115967873 | 1.40 |
ENSMUST00000153408.8
|
Unc13d
|
unc-13 homolog D |
| chr10_-_129000620 | 1.37 |
ENSMUST00000214271.2
|
Olfr771
|
olfactory receptor 771 |
| chr4_-_137137088 | 1.36 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr2_-_25124240 | 1.35 |
ENSMUST00000006638.8
|
Slc34a3
|
solute carrier family 34 (sodium phosphate), member 3 |
| chrX_-_133177717 | 1.35 |
ENSMUST00000087541.12
ENSMUST00000087540.4 |
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr4_+_52964547 | 1.32 |
ENSMUST00000215010.2
ENSMUST00000215127.2 |
Olfr270
|
olfactory receptor 270 |
| chrX_-_73067351 | 1.32 |
ENSMUST00000114353.10
ENSMUST00000101458.9 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr15_+_95698574 | 1.31 |
ENSMUST00000226793.2
|
Ano6
|
anoctamin 6 |
| chr11_-_101442663 | 1.30 |
ENSMUST00000017290.11
|
Brca1
|
breast cancer 1, early onset |
| chr4_-_4077510 | 1.29 |
ENSMUST00000108383.2
|
Sdr16c6
|
short chain dehydrogenase/reductase family 16C, member 6 |
| chrX_-_133012457 | 1.28 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chr5_-_83502793 | 1.27 |
ENSMUST00000146669.2
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr11_+_61544085 | 1.26 |
ENSMUST00000004959.3
|
Grap
|
GRB2-related adaptor protein |
| chr9_-_50439942 | 1.25 |
ENSMUST00000214873.2
ENSMUST00000034570.7 ENSMUST00000213144.2 ENSMUST00000215416.2 |
Pts
|
6-pyruvoyl-tetrahydropterin synthase |
| chr4_-_3973586 | 1.23 |
ENSMUST00000089430.6
|
Gm11808
|
predicted gene 11808 |
| chr4_-_150087587 | 1.21 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr2_+_24181208 | 1.20 |
ENSMUST00000058056.2
|
Il1f10
|
interleukin 1 family, member 10 |
| chr13_+_19376428 | 1.19 |
ENSMUST00000199017.2
|
Trgv5
|
T cell receptor gamma, variable 5 |
| chrX_-_133177638 | 1.18 |
ENSMUST00000113252.8
|
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr15_-_60696790 | 1.18 |
ENSMUST00000100635.5
|
Lratd2
|
LRAT domain containing 1 |
| chr9_+_19047613 | 1.18 |
ENSMUST00000215699.2
|
Olfr837
|
olfactory receptor 837 |
| chrX_-_73067514 | 1.15 |
ENSMUST00000033769.15
ENSMUST00000114352.8 ENSMUST00000068286.12 ENSMUST00000114360.10 ENSMUST00000114354.10 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr7_+_27196760 | 1.14 |
ENSMUST00000238964.2
|
Prx
|
periaxin |
| chr7_+_17605684 | 1.12 |
ENSMUST00000072381.14
|
Gm5155
|
predicted gene 5155 |
| chr1_+_172204109 | 1.10 |
ENSMUST00000052455.4
|
Pigm
|
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr5_+_42225303 | 1.08 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr15_+_89206923 | 1.07 |
ENSMUST00000066991.7
|
Adm2
|
adrenomedullin 2 |
| chr13_-_40441487 | 1.07 |
ENSMUST00000054635.8
|
Ofcc1
|
orofacial cleft 1 candidate 1 |
| chr6_-_40562700 | 1.03 |
ENSMUST00000177178.2
ENSMUST00000129948.9 ENSMUST00000101491.11 |
Clec5a
|
C-type lectin domain family 5, member a |
| chr16_-_50252703 | 1.02 |
ENSMUST00000066037.13
ENSMUST00000089399.11 ENSMUST00000089404.10 ENSMUST00000138166.8 |
Bbx
|
bobby sox HMG box containing |
| chr4_+_15265798 | 1.00 |
ENSMUST00000062684.9
|
Tmem64
|
transmembrane protein 64 |
| chr13_-_12814957 | 1.00 |
ENSMUST00000222139.2
ENSMUST00000099805.3 |
Prl2c3
|
prolactin family 2, subfamily c, member 3 |
| chr14_+_40853734 | 1.00 |
ENSMUST00000022314.10
ENSMUST00000170719.2 |
Sftpa1
|
surfactant associated protein A1 |
| chr13_+_19524136 | 0.99 |
ENSMUST00000103564.3
|
Trgv1
|
T cell receptor gamma, variable 1 |
| chr2_-_73283010 | 0.98 |
ENSMUST00000151939.2
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr6_+_43149074 | 0.98 |
ENSMUST00000216179.2
|
Olfr13
|
olfactory receptor 13 |
| chr13_+_19376603 | 0.97 |
ENSMUST00000103556.3
|
Trgv5
|
T cell receptor gamma, variable 5 |
| chr19_-_10655391 | 0.97 |
ENSMUST00000025647.7
|
Pga5
|
pepsinogen 5, group I |
| chr13_+_13357291 | 0.96 |
ENSMUST00000021778.14
ENSMUST00000126540.8 ENSMUST00000151144.2 |
Prl2c5
|
prolactin family 2, subfamily c, member 5 |
| chr9_-_122016671 | 0.95 |
ENSMUST00000216738.2
|
Ano10
|
anoctamin 10 |
| chr9_+_123901979 | 0.95 |
ENSMUST00000171719.8
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr2_+_72306503 | 0.93 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr8_+_85428391 | 0.92 |
ENSMUST00000238338.2
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr19_+_24853039 | 0.92 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr16_-_22848153 | 0.92 |
ENSMUST00000232459.2
|
Kng2
|
kininogen 2 |
| chr13_-_27900716 | 0.89 |
ENSMUST00000224026.2
ENSMUST00000021776.4 |
Prl7d1
|
prolactin family 7, subfamily d, member 1 |
| chr17_-_46956054 | 0.86 |
ENSMUST00000003642.7
|
Klc4
|
kinesin light chain 4 |
| chr2_-_164080358 | 0.86 |
ENSMUST00000044953.3
|
Svs2
|
seminal vesicle secretory protein 2 |
| chr7_-_103502404 | 0.86 |
ENSMUST00000033229.5
|
Hbb-y
|
hemoglobin Y, beta-like embryonic chain |
| chr13_-_67480588 | 0.83 |
ENSMUST00000109735.9
ENSMUST00000168892.9 |
Zfp595
|
zinc finger protein 595 |
| chr19_-_11838430 | 0.83 |
ENSMUST00000214796.2
|
Olfr1418
|
olfactory receptor 1418 |
| chr5_+_104318542 | 0.80 |
ENSMUST00000112771.2
|
Dspp
|
dentin sialophosphoprotein |
| chr3_+_93052089 | 0.79 |
ENSMUST00000107300.7
ENSMUST00000195515.2 |
Crnn
|
cornulin |
| chr3_-_34135408 | 0.73 |
ENSMUST00000117223.4
ENSMUST00000120805.8 ENSMUST00000011029.12 ENSMUST00000108195.10 |
Dnajc19
|
DnaJ heat shock protein family (Hsp40) member C19 |
| chr7_+_13011180 | 0.73 |
ENSMUST00000177588.10
|
Lig1
|
ligase I, DNA, ATP-dependent |
| chrX_+_52076998 | 0.72 |
ENSMUST00000026723.9
|
Hprt
|
hypoxanthine guanine phosphoribosyl transferase |
| chr14_+_53691753 | 0.72 |
ENSMUST00000180711.3
ENSMUST00000184650.2 |
Trav6-4
|
T cell receptor alpha variable 6-4 |
| chr1_+_180770064 | 0.71 |
ENSMUST00000159436.8
|
Tmem63a
|
transmembrane protein 63a |
| chr11_-_8900539 | 0.71 |
ENSMUST00000178195.2
|
Pkd1l1
|
polycystic kidney disease 1 like 1 |
| chr16_-_33916354 | 0.71 |
ENSMUST00000114973.9
ENSMUST00000232157.2 ENSMUST00000114964.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chrX_+_73636285 | 0.70 |
ENSMUST00000216108.2
ENSMUST00000216007.2 |
Olfr1325
|
olfactory receptor 1325 |
| chr5_-_83502966 | 0.70 |
ENSMUST00000053543.11
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr7_-_30555592 | 0.69 |
ENSMUST00000185748.2
ENSMUST00000094583.2 |
Ffar3
|
free fatty acid receptor 3 |
| chr9_-_35030479 | 0.69 |
ENSMUST00000213526.2
ENSMUST00000215089.2 |
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chrX_+_139808351 | 0.68 |
ENSMUST00000033806.5
|
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr5_-_113968483 | 0.67 |
ENSMUST00000100874.6
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr13_-_62519750 | 0.65 |
ENSMUST00000107989.7
|
Gm3604
|
predicted gene 3604 |
| chr10_+_127595590 | 0.64 |
ENSMUST00000073639.6
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr16_-_58850641 | 0.64 |
ENSMUST00000216415.2
ENSMUST00000206463.3 |
Olfr186
|
olfactory receptor 186 |
| chr13_-_67480562 | 0.61 |
ENSMUST00000171466.2
|
Zfp595
|
zinc finger protein 595 |
| chr1_+_185187000 | 0.61 |
ENSMUST00000061093.7
|
Slc30a10
|
solute carrier family 30, member 10 |
| chr8_+_53964721 | 0.60 |
ENSMUST00000211424.2
ENSMUST00000033920.6 |
Aga
|
aspartylglucosaminidase |
| chr13_-_62668265 | 0.59 |
ENSMUST00000082203.7
|
Zfp934
|
zinc finger protein 934 |
| chr4_-_143383816 | 0.58 |
ENSMUST00000105771.2
|
Pramel22
|
PRAME like 22 |
| chr17_-_50401305 | 0.58 |
ENSMUST00000113195.8
|
Rftn1
|
raftlin lipid raft linker 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:0002215 | defense response to nematode(GO:0002215) |
| 3.4 | 10.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 2.7 | 8.0 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 2.3 | 9.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.7 | 5.1 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 1.6 | 6.3 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 1.3 | 5.2 | GO:0045575 | basophil activation(GO:0045575) |
| 1.2 | 3.7 | GO:0035874 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 1.2 | 5.9 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 1.1 | 6.5 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.9 | 11.6 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.8 | 3.2 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.8 | 5.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.7 | 10.7 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.6 | 3.2 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.6 | 3.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.6 | 8.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.6 | 3.4 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) extrathymic T cell selection(GO:0045062) cellular response to interleukin-15(GO:0071350) |
| 0.5 | 5.4 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.5 | 4.4 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.5 | 1.4 | GO:1904826 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.4 | 1.3 | GO:0097045 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.4 | 1.7 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.4 | 2.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.4 | 4.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.4 | 20.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.4 | 3.9 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.3 | 1.0 | GO:0090264 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.3 | 6.8 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.3 | 2.5 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 14.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 2.0 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.3 | 3.3 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 0.8 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) cellular response to cell-matrix adhesion(GO:0071460) |
| 0.3 | 1.3 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.3 | 3.8 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 1.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 1.7 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 0.7 | GO:0046101 | GMP catabolic process(GO:0046038) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.2 | 0.7 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.2 | 1.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.2 | 1.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.2 | 2.7 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 13.4 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.2 | 4.5 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.2 | 0.6 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.2 | 3.9 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.2 | 2.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 2.0 | GO:0070627 | ferrous iron import(GO:0070627) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.2 | 6.9 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.2 | 1.4 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.2 | 0.7 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.2 | 2.8 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.2 | 2.6 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.2 | 2.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 1.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 0.6 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.7 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.7 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 0.6 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 1.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 3.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 2.6 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 0.4 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.7 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.1 | 0.7 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 1.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 2.0 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 4.0 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 1.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 4.9 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.1 | 7.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 3.5 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 9.7 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 4.8 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 0.9 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.0 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 4.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 9.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 1.4 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.0 | 0.3 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 4.2 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 2.4 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 5.5 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 | 0.9 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 1.0 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 4.3 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.3 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.7 | GO:0048806 | genitalia development(GO:0048806) |
| 0.0 | 5.7 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 1.7 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.6 | GO:0001523 | retinoid metabolic process(GO:0001523) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 22.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.2 | 8.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.6 | 3.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.6 | 1.7 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.4 | 5.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 3.0 | GO:1990462 | omegasome(GO:1990462) |
| 0.2 | 8.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 2.6 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.2 | 2.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 4.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 4.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 4.3 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 10.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 2.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 4.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 1.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 5.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 11.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 1.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 0.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 7.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 6.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 3.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 4.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 3.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 11.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 5.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 19.5 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 7.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 2.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 63.3 | GO:0044421 | extracellular region part(GO:0044421) |
| 0.0 | 2.0 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.7 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 14.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 5.4 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 22.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 2.1 | 6.3 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 2.0 | 20.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.8 | 12.3 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 1.7 | 10.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 1.7 | 5.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 1.2 | 3.7 | GO:0052600 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 1.1 | 8.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 1.1 | 3.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 1.1 | 3.2 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 1.1 | 6.3 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.9 | 2.6 | GO:0019002 | GMP binding(GO:0019002) |
| 0.8 | 4.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.8 | 4.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.7 | 5.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.6 | 5.5 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.6 | 1.7 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.5 | 6.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.4 | 0.9 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.4 | 2.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.4 | 1.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.4 | 1.2 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.4 | 2.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.4 | 14.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.4 | 9.4 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.3 | 5.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.3 | 2.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 1.0 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.3 | 3.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.3 | 1.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.3 | 3.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 3.9 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 3.8 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 10.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 7.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.4 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.2 | 4.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 3.0 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.6 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 8.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 3.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 4.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 6.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.7 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 2.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.7 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.5 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 20.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 7.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 27.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 2.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 4.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.7 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 3.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 3.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 3.9 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 1.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.1 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 2.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 2.1 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 0.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 3.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 4.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 4.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.0 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 5.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 4.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.0 | 1.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 2.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 3.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 2.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.7 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 4.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 14.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 15.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 14.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 4.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 2.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 6.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 7.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 4.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 5.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 7.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 11.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.5 | PID ENDOTHELIN PATHWAY | Endothelins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.8 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.4 | 17.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.3 | 3.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 6.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 3.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 14.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 6.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.2 | 3.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 4.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 5.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.2 | 2.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 3.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 4.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 5.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 5.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 3.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 2.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 13.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 4.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 6.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 3.3 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |