Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
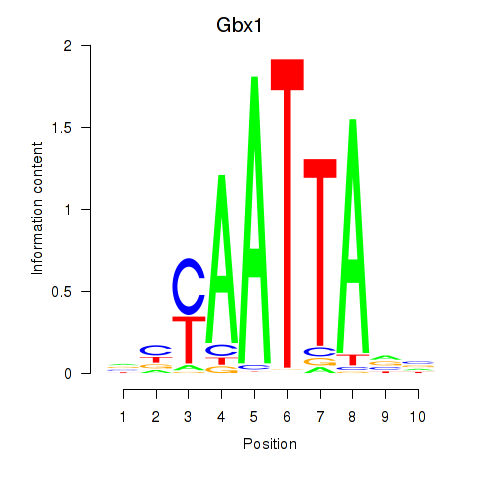
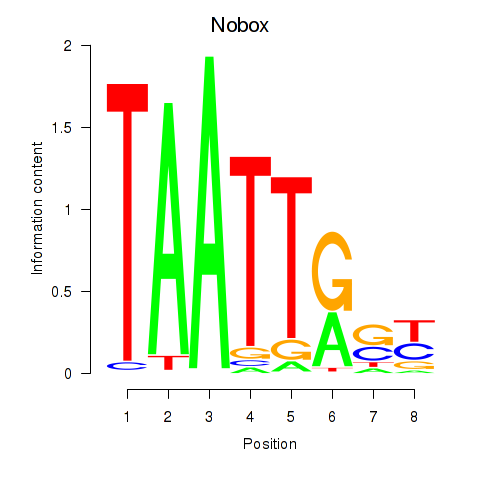
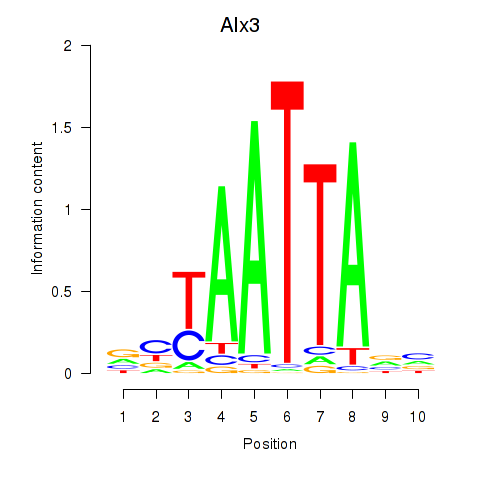
Results for Gbx1_Nobox_Alx3
Z-value: 0.84
Transcription factors associated with Gbx1_Nobox_Alx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gbx1
|
ENSMUSG00000067724.6 | Gbx1 |
|
Nobox
|
ENSMUSG00000029736.16 | Nobox |
|
Alx3
|
ENSMUSG00000014603.4 | Alx3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Alx3 | mm39_v1_chr3_+_107502347_107502479 | 0.58 | 7.9e-08 | Click! |
| Gbx1 | mm39_v1_chr5_-_24732200_24732200 | 0.42 | 2.1e-04 | Click! |
| Nobox | mm39_v1_chr6_-_43284170_43284170 | 0.02 | 8.9e-01 | Click! |
Activity profile of Gbx1_Nobox_Alx3 motif
Sorted Z-values of Gbx1_Nobox_Alx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gbx1_Nobox_Alx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 1.5 | 9.0 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.2 | 4.9 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 1.1 | 7.8 | GO:0061743 | motor learning(GO:0061743) |
| 1.1 | 3.3 | GO:0015825 | L-serine transport(GO:0015825) |
| 1.0 | 12.7 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.9 | 17.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.8 | 5.0 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.8 | 4.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.8 | 2.4 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.8 | 3.1 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.7 | 2.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.7 | 3.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.7 | 4.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.7 | 10.6 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.6 | 3.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.6 | 2.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.6 | 6.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.5 | 3.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.5 | 3.7 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.5 | 1.5 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.5 | 1.0 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.5 | 2.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.5 | 1.8 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.4 | 1.8 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.4 | 2.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.4 | 1.2 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.4 | 7.0 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.4 | 2.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.4 | 3.2 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.4 | 14.7 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.4 | 1.8 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.4 | 1.4 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.4 | 1.4 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.3 | 2.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.3 | 1.0 | GO:1990751 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.3 | 5.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.3 | 4.6 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.3 | 2.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.3 | 1.9 | GO:0060335 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) heme transport(GO:0015886) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 8.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.3 | 2.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.2 | 1.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 1.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 2.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 1.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 1.7 | GO:0060287 | determination of pancreatic left/right asymmetry(GO:0035469) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 10.4 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.2 | 1.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.2 | 2.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 1.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.2 | 0.4 | GO:2000292 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.2 | 0.8 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 1.4 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.2 | 0.5 | GO:0060618 | nipple development(GO:0060618) |
| 0.2 | 0.7 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.2 | 0.5 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.2 | 9.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.2 | 1.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 1.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.0 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 1.0 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.2 | 0.9 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.2 | 1.6 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 5.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 6.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 21.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 5.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 3.0 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.1 | 3.4 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 3.9 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 1.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.4 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 1.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 3.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.8 | GO:1904936 | forebrain anterior/posterior pattern specification(GO:0021797) cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.6 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 1.1 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.2 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 1.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 2.3 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 0.5 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.6 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 1.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.6 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 4.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 4.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.5 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 | 1.5 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 6.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 8.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.4 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 2.6 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 10.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 2.0 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 | 1.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 1.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 1.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 2.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 4.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 2.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.6 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.1 | 0.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.4 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 1.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 1.0 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 0.4 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 0.5 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 2.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.6 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 5.8 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 6.0 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.4 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 1.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 1.5 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 1.8 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.6 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 1.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.0 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.5 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 1.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 4.5 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 1.0 | GO:0060444 | branching involved in mammary gland duct morphogenesis(GO:0060444) |
| 0.0 | 2.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 1.1 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.7 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 1.9 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 1.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 1.6 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 1.5 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 1.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.4 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.4 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 1.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 1.9 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.4 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 2.0 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.4 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 1.0 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 1.5 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 1.7 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.2 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 1.3 | 5.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.7 | 2.1 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.5 | 2.6 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.5 | 9.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 0.4 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.4 | 1.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.4 | 4.0 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.4 | 2.3 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.4 | 6.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 8.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.3 | 12.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 2.6 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.3 | 3.3 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 1.6 | GO:0071547 | piP-body(GO:0071547) |
| 0.3 | 1.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 1.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.2 | 0.6 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.2 | 4.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 3.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.0 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.7 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 4.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 2.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 2.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.4 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 1.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 4.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 10.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 4.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.4 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.1 | 4.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 4.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 3.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 2.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 9.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 5.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 2.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 11.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 34.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 1.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 7.0 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 6.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 4.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 2.0 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 6.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 1.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 8.2 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 2.6 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.8 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 10.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.7 | 5.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.6 | 3.1 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.6 | 3.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.6 | 10.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.6 | 4.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.5 | 4.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.5 | 20.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.5 | 1.9 | GO:0019862 | IgA binding(GO:0019862) |
| 0.5 | 3.2 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.5 | 2.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 2.7 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.4 | 1.4 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.4 | 5.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.3 | 3.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 2.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 3.3 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.3 | 5.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 1.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 10.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.3 | 1.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 1.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.3 | 2.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.3 | 2.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 1.1 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.3 | 1.0 | GO:0051381 | histamine binding(GO:0051381) |
| 0.2 | 1.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 1.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 4.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 3.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 2.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 1.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 2.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 1.0 | GO:0015100 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 5.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 0.9 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 5.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 8.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 5.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 5.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.5 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.7 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 3.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 1.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 2.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 4.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 17.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.5 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 12.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 13.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.9 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.5 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 2.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 1.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 2.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 5.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 7.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 13.3 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 3.2 | GO:0017016 | Ras GTPase binding(GO:0017016) |
| 0.0 | 26.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 1.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 2.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 8.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 6.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 2.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 1.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 3.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.8 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 18.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 10.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 7.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.8 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 5.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 6.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 7.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.7 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 0.8 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.4 | 10.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 11.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 5.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 3.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 2.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 5.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 2.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 6.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 4.0 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 4.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 4.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 3.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 8.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 0.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 1.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 3.0 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 4.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.9 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.9 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |