Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
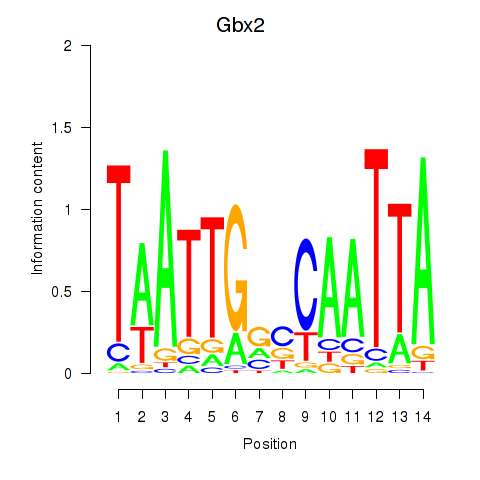
Results for Gbx2
Z-value: 0.77
Transcription factors associated with Gbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gbx2
|
ENSMUSG00000034486.9 | Gbx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gbx2 | mm39_v1_chr1_-_89858901_89858920 | 0.36 | 2.0e-03 | Click! |
Activity profile of Gbx2 motif
Sorted Z-values of Gbx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gbx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_79394904 | 10.37 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr2_+_70392351 | 7.86 |
ENSMUST00000094934.11
|
Gad1
|
glutamate decarboxylase 1 |
| chr2_+_70392491 | 6.18 |
ENSMUST00000148210.8
|
Gad1
|
glutamate decarboxylase 1 |
| chr14_-_9015639 | 6.06 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chr12_+_74044435 | 6.05 |
ENSMUST00000221220.2
|
Syt16
|
synaptotagmin XVI |
| chr7_-_141014477 | 5.87 |
ENSMUST00000106007.10
ENSMUST00000150026.2 ENSMUST00000202840.4 ENSMUST00000133206.9 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr19_+_17114108 | 5.67 |
ENSMUST00000223920.2
ENSMUST00000225351.2 |
Prune2
|
prune homolog 2 |
| chr7_-_141014445 | 5.47 |
ENSMUST00000133021.2
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr9_+_108708939 | 5.18 |
ENSMUST00000192235.2
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr7_-_141014192 | 5.17 |
ENSMUST00000201127.5
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr7_-_141014336 | 5.09 |
ENSMUST00000136354.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr7_-_140402037 | 4.72 |
ENSMUST00000106052.2
ENSMUST00000080651.13 |
Zfp941
|
zinc finger protein 941 |
| chrX_-_42256694 | 4.72 |
ENSMUST00000115058.8
ENSMUST00000115059.8 |
Tenm1
|
teneurin transmembrane protein 1 |
| chr10_-_5144699 | 4.35 |
ENSMUST00000215467.2
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr6_+_114435480 | 4.29 |
ENSMUST00000160780.2
|
Hrh1
|
histamine receptor H1 |
| chr7_-_105230395 | 4.25 |
ENSMUST00000188726.2
ENSMUST00000188440.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr7_-_105230807 | 4.25 |
ENSMUST00000191011.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr7_-_105230698 | 4.16 |
ENSMUST00000189378.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr4_+_110254858 | 4.14 |
ENSMUST00000106589.9
ENSMUST00000106587.9 ENSMUST00000106591.8 ENSMUST00000106592.8 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr14_-_9015757 | 3.66 |
ENSMUST00000153954.8
|
Synpr
|
synaptoporin |
| chr18_+_37125424 | 3.47 |
ENSMUST00000194038.2
|
Pcdha8
|
protocadherin alpha 8 |
| chr10_+_116013256 | 3.15 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr5_-_5315968 | 3.03 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr5_-_31453206 | 2.95 |
ENSMUST00000041266.11
ENSMUST00000172435.8 ENSMUST00000201417.2 |
Fndc4
|
fibronectin type III domain containing 4 |
| chr3_-_113371392 | 2.83 |
ENSMUST00000067980.12
|
Amy1
|
amylase 1, salivary |
| chr4_+_110254907 | 2.72 |
ENSMUST00000097920.9
ENSMUST00000080744.13 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr10_+_116013122 | 2.63 |
ENSMUST00000148731.8
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr5_+_117919082 | 2.36 |
ENSMUST00000138579.3
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr1_+_179936757 | 2.13 |
ENSMUST00000143176.8
ENSMUST00000135056.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_-_17465410 | 2.11 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chrM_+_14138 | 2.04 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr17_-_33028814 | 1.99 |
ENSMUST00000234594.2
|
Zfp811
|
zinc finger protein 811 |
| chr14_-_66451001 | 1.99 |
ENSMUST00000178730.8
|
Ptk2b
|
PTK2 protein tyrosine kinase 2 beta |
| chr2_-_157408239 | 1.98 |
ENSMUST00000109528.9
ENSMUST00000088494.3 |
Blcap
|
bladder cancer associated protein |
| chr18_+_77877611 | 1.69 |
ENSMUST00000238172.2
|
Pstpip2
|
proline-serine-threonine phosphatase-interacting protein 2 |
| chr16_-_36605286 | 1.61 |
ENSMUST00000168279.2
ENSMUST00000164579.9 ENSMUST00000023616.11 |
Slc15a2
|
solute carrier family 15 (H+/peptide transporter), member 2 |
| chr14_-_48904958 | 1.56 |
ENSMUST00000144465.8
ENSMUST00000133479.8 ENSMUST00000119070.8 ENSMUST00000226501.2 |
Otx2
|
orthodenticle homeobox 2 |
| chr4_-_139695337 | 1.55 |
ENSMUST00000105031.4
|
Klhdc7a
|
kelch domain containing 7A |
| chr3_-_86827664 | 1.52 |
ENSMUST00000194452.2
ENSMUST00000191752.6 |
Dclk2
|
doublecortin-like kinase 2 |
| chr16_-_36605147 | 1.51 |
ENSMUST00000165531.9
|
Slc15a2
|
solute carrier family 15 (H+/peptide transporter), member 2 |
| chr2_-_18806629 | 1.43 |
ENSMUST00000181515.2
|
Carlr
|
cardiac and apoptosis-related long non-coding RNA |
| chr5_+_113873864 | 1.38 |
ENSMUST00000065698.7
|
Ficd
|
FIC domain containing |
| chr4_-_26346882 | 1.35 |
ENSMUST00000041374.8
ENSMUST00000153813.2 |
Manea
|
mannosidase, endo-alpha |
| chr19_-_39729431 | 1.26 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr3_-_141687987 | 1.09 |
ENSMUST00000029948.15
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr11_+_59197746 | 1.03 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr14_+_69585036 | 1.03 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chrM_-_14061 | 1.03 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr1_-_52991993 | 1.02 |
ENSMUST00000050567.11
|
1700019D03Rik
|
RIKEN cDNA 1700019D03 gene |
| chr3_-_86827640 | 1.02 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr2_+_87609827 | 1.02 |
ENSMUST00000105210.3
|
Olfr152
|
olfactory receptor 152 |
| chr18_-_67378886 | 0.97 |
ENSMUST00000073054.5
|
Mppe1
|
metallophosphoesterase 1 |
| chr11_+_97206542 | 0.95 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr1_-_155293141 | 0.92 |
ENSMUST00000111775.8
ENSMUST00000111774.2 |
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr11_-_21320452 | 0.83 |
ENSMUST00000102875.11
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr2_-_89675315 | 0.79 |
ENSMUST00000099762.2
|
Olfr48
|
olfactory receptor 48 |
| chr17_-_37974666 | 0.69 |
ENSMUST00000215414.2
ENSMUST00000213638.3 |
Olfr117
|
olfactory receptor 117 |
| chr2_-_73410632 | 0.68 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr4_+_118818775 | 0.65 |
ENSMUST00000058651.5
|
Lao1
|
L-amino acid oxidase 1 |
| chr10_-_95337783 | 0.64 |
ENSMUST00000075829.3
ENSMUST00000217777.2 ENSMUST00000218893.2 |
Mrpl42
|
mitochondrial ribosomal protein L42 |
| chr3_-_89905927 | 0.61 |
ENSMUST00000197725.5
ENSMUST00000197767.5 ENSMUST00000197786.5 ENSMUST00000079724.9 |
Hax1
|
HCLS1 associated X-1 |
| chr4_+_32657105 | 0.61 |
ENSMUST00000071642.11
ENSMUST00000178134.2 |
Mdn1
|
midasin AAA ATPase 1 |
| chr1_-_74932266 | 0.58 |
ENSMUST00000006721.3
|
Cryba2
|
crystallin, beta A2 |
| chr19_-_12313274 | 0.56 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1 |
| chr2_-_85966272 | 0.54 |
ENSMUST00000216566.3
ENSMUST00000214364.2 |
Olfr1039
|
olfactory receptor 1039 |
| chr10_-_23112973 | 0.53 |
ENSMUST00000218049.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr17_+_85264134 | 0.51 |
ENSMUST00000112305.10
|
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr6_-_129077867 | 0.50 |
ENSMUST00000032258.8
|
Clec2e
|
C-type lectin domain family 2, member e |
| chr10_-_128918779 | 0.42 |
ENSMUST00000213579.2
|
Olfr767
|
olfactory receptor 767 |
| chr8_+_72837021 | 0.41 |
ENSMUST00000213940.2
|
Olfr373
|
olfactory receptor 373 |
| chr14_+_54418031 | 0.39 |
ENSMUST00000103704.2
|
Traj38
|
T cell receptor alpha joining 38 |
| chr4_+_114020581 | 0.37 |
ENSMUST00000079915.10
ENSMUST00000164297.8 |
Skint11
|
selection and upkeep of intraepithelial T cells 11 |
| chr3_+_88523440 | 0.37 |
ENSMUST00000177498.8
ENSMUST00000176500.8 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr7_-_78432774 | 0.34 |
ENSMUST00000032841.7
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr2_-_87838612 | 0.33 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160 |
| chr3_-_10396418 | 0.33 |
ENSMUST00000191670.6
ENSMUST00000065938.15 ENSMUST00000118410.8 |
Impa1
|
inositol (myo)-1(or 4)-monophosphatase 1 |
| chr10_-_116732813 | 0.32 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr7_+_78432867 | 0.27 |
ENSMUST00000032840.5
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr2_+_67004178 | 0.23 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr2_-_111324108 | 0.19 |
ENSMUST00000208881.2
ENSMUST00000208695.2 ENSMUST00000217611.2 |
Olfr1290
|
olfactory receptor 1290 |
| chr4_+_114020656 | 0.17 |
ENSMUST00000145797.8
ENSMUST00000151810.2 |
Skint11
|
selection and upkeep of intraepithelial T cells 11 |
| chr14_+_54436898 | 0.17 |
ENSMUST00000103721.3
|
Traj20
|
T cell receptor alpha joining 20 |
| chr9_+_37656402 | 0.10 |
ENSMUST00000216982.2
|
Olfr874
|
olfactory receptor 874 |
| chr14_-_50390356 | 0.08 |
ENSMUST00000215451.2
ENSMUST00000213163.2 ENSMUST00000215327.2 |
Olfr729
|
olfactory receptor 729 |
| chr12_-_98225676 | 0.03 |
ENSMUST00000021390.9
|
Galc
|
galactosylceramidase |
| chr14_-_48904998 | 0.00 |
ENSMUST00000152018.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr11_-_73707875 | 0.00 |
ENSMUST00000118463.4
ENSMUST00000144724.3 |
Olfr392
|
olfactory receptor 392 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.7 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 2.0 | 14.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.4 | 6.9 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 1.0 | 5.2 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 1.0 | 5.8 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.7 | 4.3 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.7 | 21.6 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.7 | 2.0 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.6 | 4.3 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.6 | 2.4 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.4 | 3.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.4 | 10.4 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.3 | 0.8 | GO:0019255 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.3 | 2.0 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.2 | 1.6 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.2 | 1.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 1.4 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 | 2.5 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 2.1 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.5 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.9 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) phosphate ion transmembrane transport(GO:0035435) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 4.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.0 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.7 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.6 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.5 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 1.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 6.0 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 2.8 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.0 | 2.2 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 12.7 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.6 | 3.0 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.4 | 4.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.3 | 9.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 14.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 1.0 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.3 | 1.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 2.0 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 2.4 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 2.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 6.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 9.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 21.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.4 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 14.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 1.3 | 21.6 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 1.1 | 4.3 | GO:0051381 | histamine binding(GO:0051381) |
| 1.0 | 3.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.7 | 2.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.5 | 2.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.4 | 2.8 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.4 | 12.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 2.0 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 0.9 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 6.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 0.7 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.2 | 1.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 4.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.8 | GO:0070569 | pyrimidine ribonucleotide binding(GO:0032557) uridylyltransferase activity(GO:0070569) |
| 0.1 | 1.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 6.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.3 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 3.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 1.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 6.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.7 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 1.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.8 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 14.8 | GO:0042803 | protein homodimerization activity(GO:0042803) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 12.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 10.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 2.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 14.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 4.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 4.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 10.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 2.4 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 2.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 3.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |