Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Gcm1
Z-value: 0.56
Transcription factors associated with Gcm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gcm1
|
ENSMUSG00000023333.9 | Gcm1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gcm1 | mm39_v1_chr9_+_77959206_77959231 | 0.07 | 5.7e-01 | Click! |
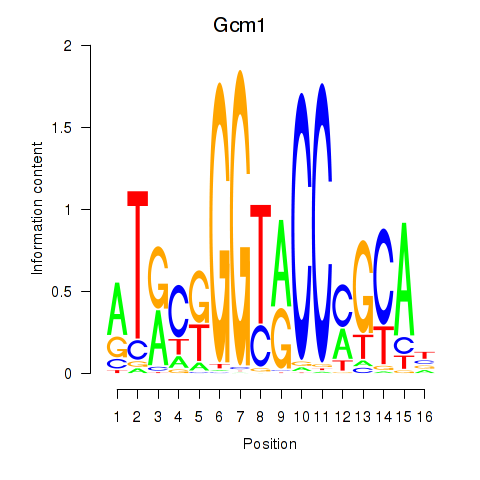
Activity profile of Gcm1 motif
Sorted Z-values of Gcm1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gcm1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_68013772 | 3.38 |
ENSMUST00000197515.2
ENSMUST00000103315.3 |
Igkv17-121
|
immunoglobulin kappa variable 17-121 |
| chr7_-_100661181 | 3.28 |
ENSMUST00000178340.3
ENSMUST00000037540.5 |
P2ry2
|
purinergic receptor P2Y, G-protein coupled 2 |
| chr8_-_3770642 | 3.00 |
ENSMUST00000062037.7
|
Clec4g
|
C-type lectin domain family 4, member g |
| chr6_-_70318437 | 2.88 |
ENSMUST00000196599.2
|
Igkv8-19
|
immunoglobulin kappa variable 8-19 |
| chr11_+_101137231 | 2.58 |
ENSMUST00000122006.8
ENSMUST00000151830.2 |
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr6_-_70120881 | 2.57 |
ENSMUST00000103380.3
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chr11_+_101137786 | 2.56 |
ENSMUST00000107282.4
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr7_-_100661220 | 2.50 |
ENSMUST00000207916.2
|
P2ry2
|
purinergic receptor P2Y, G-protein coupled 2 |
| chr2_-_34951443 | 2.37 |
ENSMUST00000028233.7
|
Hc
|
hemolytic complement |
| chr17_-_32408431 | 2.30 |
ENSMUST00000087721.10
ENSMUST00000162117.3 |
Ephx3
|
epoxide hydrolase 3 |
| chr19_+_6450553 | 2.30 |
ENSMUST00000146831.8
ENSMUST00000035716.15 ENSMUST00000138555.8 ENSMUST00000167240.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr8_-_13304154 | 2.21 |
ENSMUST00000204916.3
ENSMUST00000033825.11 |
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr6_-_124698805 | 2.11 |
ENSMUST00000173315.8
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chrX_-_74174608 | 2.05 |
ENSMUST00000033775.9
|
Mpp1
|
membrane protein, palmitoylated |
| chr16_+_65612083 | 2.01 |
ENSMUST00000168064.3
|
Vgll3
|
vestigial like family member 3 |
| chrX_-_74174524 | 1.97 |
ENSMUST00000114091.8
|
Mpp1
|
membrane protein, palmitoylated |
| chr7_-_44785480 | 1.97 |
ENSMUST00000211246.2
ENSMUST00000210197.2 |
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr1_-_36578439 | 1.96 |
ENSMUST00000191849.6
|
Gm42417
|
predicted gene, 42417 |
| chr19_+_6450641 | 1.88 |
ENSMUST00000113467.2
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr8_-_13304096 | 1.87 |
ENSMUST00000171619.2
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr16_+_65612394 | 1.83 |
ENSMUST00000227997.2
|
Vgll3
|
vestigial like family member 3 |
| chr8_-_13304068 | 1.81 |
ENSMUST00000168498.8
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr17_+_23898633 | 1.71 |
ENSMUST00000233364.2
|
Cldn6
|
claudin 6 |
| chr18_-_39620115 | 1.70 |
ENSMUST00000097592.9
ENSMUST00000115571.8 |
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr7_-_44785815 | 1.68 |
ENSMUST00000146760.7
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr7_-_30560989 | 1.67 |
ENSMUST00000052700.6
|
Ffar1
|
free fatty acid receptor 1 |
| chr6_+_67532481 | 1.60 |
ENSMUST00000103302.3
|
Igkv2-137
|
immunoglobulin kappa chain variable 2-137 |
| chr10_-_119948890 | 1.58 |
ENSMUST00000020449.12
|
Helb
|
helicase (DNA) B |
| chr17_+_64907697 | 1.47 |
ENSMUST00000086723.10
|
Man2a1
|
mannosidase 2, alpha 1 |
| chr3_+_90173813 | 1.44 |
ENSMUST00000098914.10
|
Dennd4b
|
DENN/MADD domain containing 4B |
| chr12_-_73160181 | 1.44 |
ENSMUST00000043208.8
ENSMUST00000175693.3 |
Six4
|
sine oculis-related homeobox 4 |
| chr17_+_23898223 | 1.41 |
ENSMUST00000024699.4
ENSMUST00000232719.2 |
Cldn6
|
claudin 6 |
| chrX_+_7523499 | 1.39 |
ENSMUST00000033485.14
|
Prickle3
|
prickle planar cell polarity protein 3 |
| chr2_+_127967951 | 1.36 |
ENSMUST00000089634.12
ENSMUST00000019281.14 ENSMUST00000110341.9 ENSMUST00000103211.8 ENSMUST00000103210.2 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chr7_-_44785709 | 1.34 |
ENSMUST00000211429.2
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr6_-_69394425 | 1.34 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chr7_+_126895531 | 1.27 |
ENSMUST00000170971.8
|
Itgal
|
integrin alpha L |
| chr6_-_70313491 | 1.26 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr6_-_124613044 | 1.13 |
ENSMUST00000068797.3
ENSMUST00000218020.2 |
C1s2
|
complement component 1, s subcomponent 2 |
| chr9_-_107482462 | 1.12 |
ENSMUST00000194433.6
|
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr9_-_107482494 | 1.07 |
ENSMUST00000102529.10
|
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr13_-_4573312 | 1.07 |
ENSMUST00000221564.2
ENSMUST00000078239.5 ENSMUST00000080361.13 |
Akr1c20
|
aldo-keto reductase family 1, member C20 |
| chr16_-_36695166 | 1.07 |
ENSMUST00000075946.12
|
Eaf2
|
ELL associated factor 2 |
| chr1_+_53352780 | 0.97 |
ENSMUST00000027265.10
ENSMUST00000114484.8 |
Osgepl1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr16_-_16687119 | 0.86 |
ENSMUST00000075017.5
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr14_+_32321341 | 0.85 |
ENSMUST00000187377.7
ENSMUST00000189022.8 ENSMUST00000186452.7 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr14_+_32321824 | 0.83 |
ENSMUST00000068938.7
ENSMUST00000228878.2 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr5_+_102872838 | 0.81 |
ENSMUST00000112853.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr10_+_52265049 | 0.75 |
ENSMUST00000219730.2
|
Nepn
|
nephrocan |
| chrX_-_153999333 | 0.70 |
ENSMUST00000112551.4
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr3_+_138058139 | 0.69 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr7_+_78563513 | 0.68 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein |
| chr10_+_79984097 | 0.67 |
ENSMUST00000099492.10
ENSMUST00000042057.12 |
Midn
|
midnolin |
| chr2_+_105734975 | 0.66 |
ENSMUST00000037499.6
|
Immp1l
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr15_-_83135920 | 0.63 |
ENSMUST00000229687.2
ENSMUST00000164614.3 ENSMUST00000049530.14 |
A4galt
|
alpha 1,4-galactosyltransferase |
| chr14_+_55815879 | 0.63 |
ENSMUST00000174563.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr9_-_70564403 | 0.63 |
ENSMUST00000213380.2
ENSMUST00000049031.6 |
Mindy2
|
MINDY lysine 48 deubiquitinase 2 |
| chr6_-_84564623 | 0.62 |
ENSMUST00000205228.3
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr11_-_98040377 | 0.61 |
ENSMUST00000103143.10
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr7_-_44646960 | 0.59 |
ENSMUST00000207443.2
ENSMUST00000207755.2 ENSMUST00000003290.12 |
Bcl2l12
|
BCL2-like 12 (proline rich) |
| chr12_+_8724129 | 0.56 |
ENSMUST00000111123.9
ENSMUST00000178015.8 ENSMUST00000020915.10 |
Pum2
|
pumilio RNA-binding family member 2 |
| chr11_+_120382666 | 0.56 |
ENSMUST00000026899.4
|
Slc25a10
|
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 |
| chr14_+_55815817 | 0.56 |
ENSMUST00000174259.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr7_+_75879603 | 0.55 |
ENSMUST00000156166.8
|
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr11_-_75345482 | 0.54 |
ENSMUST00000173320.8
|
Wdr81
|
WD repeat domain 81 |
| chr9_+_107457316 | 0.52 |
ENSMUST00000093785.6
|
Naa80
|
N(alpha)-acetyltransferase 80, NatH catalytic subunit |
| chr14_-_31923803 | 0.51 |
ENSMUST00000226683.2
ENSMUST00000170331.8 ENSMUST00000013845.13 |
Timm23
|
translocase of inner mitochondrial membrane 23 |
| chr4_+_63477018 | 0.51 |
ENSMUST00000077709.11
|
Tmem268
|
transmembrane protein 268 |
| chr2_-_39116457 | 0.49 |
ENSMUST00000028087.6
|
Ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr12_+_8724220 | 0.47 |
ENSMUST00000111122.9
|
Pum2
|
pumilio RNA-binding family member 2 |
| chr14_+_55815580 | 0.45 |
ENSMUST00000174484.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr6_-_52160816 | 0.44 |
ENSMUST00000134831.2
|
Hoxa3
|
homeobox A3 |
| chr7_+_44647072 | 0.43 |
ENSMUST00000003284.16
ENSMUST00000209066.2 |
Irf3
|
interferon regulatory factor 3 |
| chr16_+_17798292 | 0.39 |
ENSMUST00000075371.5
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr6_+_86342622 | 0.37 |
ENSMUST00000071492.9
|
Fam136a
|
family with sequence similarity 136, member A |
| chr5_+_114427227 | 0.34 |
ENSMUST00000169347.6
|
Myo1h
|
myosin 1H |
| chr10_-_71121083 | 0.34 |
ENSMUST00000020085.7
|
Ube2d1
|
ubiquitin-conjugating enzyme E2D 1 |
| chr13_+_55517545 | 0.33 |
ENSMUST00000063771.14
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr3_+_107137924 | 0.33 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr10_+_75399920 | 0.31 |
ENSMUST00000141062.8
ENSMUST00000152657.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr1_-_43131598 | 0.30 |
ENSMUST00000188728.2
|
Tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr11_-_99433984 | 0.29 |
ENSMUST00000107443.8
ENSMUST00000074253.4 |
Krt40
|
keratin 40 |
| chr4_-_42661893 | 0.28 |
ENSMUST00000108006.4
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr11_-_73089837 | 0.22 |
ENSMUST00000006103.9
ENSMUST00000108476.8 |
Ctns
|
cystinosis, nephropathic |
| chr7_+_104228831 | 0.21 |
ENSMUST00000098174.3
|
Olfr653
|
olfactory receptor 653 |
| chr15_+_10249646 | 0.20 |
ENSMUST00000134410.8
|
Prlr
|
prolactin receptor |
| chr17_-_33007238 | 0.20 |
ENSMUST00000159086.10
|
Zfp871
|
zinc finger protein 871 |
| chr2_+_174257597 | 0.18 |
ENSMUST00000109075.8
ENSMUST00000016397.7 |
Nelfcd
|
negative elongation factor complex member C/D, Th1l |
| chr3_-_102689986 | 0.18 |
ENSMUST00000029450.10
ENSMUST00000200041.5 |
Tshb
|
thyroid stimulating hormone, beta subunit |
| chr3_+_89366632 | 0.17 |
ENSMUST00000107410.8
|
Pmvk
|
phosphomevalonate kinase |
| chr7_-_44785603 | 0.16 |
ENSMUST00000209467.2
|
Gm45713
|
predicted gene 45713 |
| chr3_-_102690026 | 0.15 |
ENSMUST00000172026.8
ENSMUST00000170856.6 |
Tshb
|
thyroid stimulating hormone, beta subunit |
| chr9_+_5345405 | 0.13 |
ENSMUST00000027009.11
|
Casp12
|
caspase 12 |
| chr2_-_39116284 | 0.12 |
ENSMUST00000204701.3
|
Ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr9_+_5345450 | 0.11 |
ENSMUST00000151332.2
|
Casp12
|
caspase 12 |
| chr4_-_9669068 | 0.10 |
ENSMUST00000078139.13
ENSMUST00000108340.9 ENSMUST00000084915.11 ENSMUST00000108337.8 ENSMUST00000084912.12 ENSMUST00000038564.13 ENSMUST00000146441.8 ENSMUST00000098275.9 |
Asph
|
aspartate-beta-hydroxylase |
| chr7_-_105249308 | 0.10 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr19_+_46750016 | 0.09 |
ENSMUST00000099373.12
ENSMUST00000077666.6 |
Cnnm2
|
cyclin M2 |
| chr2_-_39116044 | 0.08 |
ENSMUST00000204368.2
|
Ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr16_+_36695479 | 0.05 |
ENSMUST00000023534.7
ENSMUST00000114812.9 ENSMUST00000134616.8 |
Golgb1
|
golgi autoantigen, golgin subfamily b, macrogolgin 1 |
| chr2_-_105734829 | 0.05 |
ENSMUST00000122965.8
|
Elp4
|
elongator acetyltransferase complex subunit 4 |
| chr8_+_91796767 | 0.05 |
ENSMUST00000209518.2
|
Rbl2
|
RB transcriptional corepressor like 2 |
| chr10_-_74868360 | 0.04 |
ENSMUST00000159994.2
ENSMUST00000179546.8 ENSMUST00000160450.8 ENSMUST00000160072.8 ENSMUST00000009214.10 ENSMUST00000166088.8 |
Rsph14
|
radial spoke head homolog 14 (Chlamydomonas) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.0 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.8 | 5.9 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.6 | 5.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.5 | 5.8 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.5 | 2.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.4 | 1.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.4 | 1.4 | GO:0061055 | myotome development(GO:0061055) |
| 0.3 | 1.4 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.3 | 2.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.2 | 1.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 1.7 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 | 1.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.3 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.7 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.6 | GO:0015744 | succinate transport(GO:0015744) |
| 0.1 | 1.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 3.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.5 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.4 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.1 | 0.6 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.1 | 0.6 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 | 4.0 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.1 | 0.7 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 1.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 1.7 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.1 | 0.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 1.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.7 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.8 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 1.0 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.6 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 13.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.6 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.3 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 2.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 1.7 | GO:0010524 | positive regulation of calcium ion transport into cytosol(GO:0010524) |
| 0.0 | 0.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.1 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.4 | 1.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.3 | 1.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 2.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 1.3 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.2 | 1.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 0.7 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 2.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 5.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 1.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 3.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 4.0 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 5.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 1.5 | 5.9 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.9 | 5.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.5 | 1.5 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.3 | 1.0 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.3 | 1.7 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 1.6 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.3 | 1.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.2 | 0.6 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 0.7 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 2.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 0.6 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.1 | 1.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 2.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 3.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 3.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.7 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 5.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.5 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 1.1 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 1.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 2.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 3.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 5.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 4.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 3.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 3.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |