Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
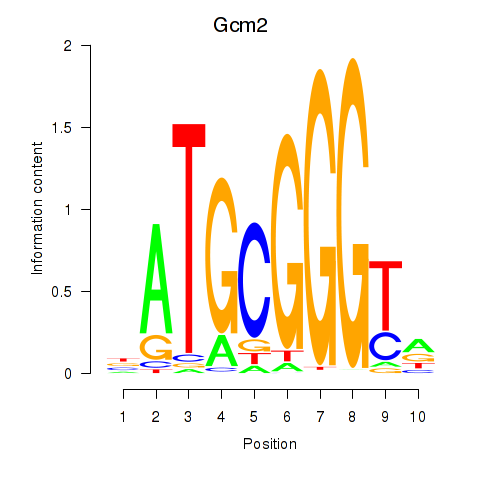
Results for Gcm2
Z-value: 1.21
Transcription factors associated with Gcm2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gcm2
|
ENSMUSG00000021362.8 | Gcm2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gcm2 | mm39_v1_chr13_-_41263659_41263659 | -0.23 | 4.8e-02 | Click! |
Activity profile of Gcm2 motif
Sorted Z-values of Gcm2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gcm2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.3 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 3.8 | 11.4 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 2.9 | 17.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 2.4 | 12.2 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 1.5 | 4.6 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 1.2 | 3.7 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 1.2 | 3.7 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 1.2 | 5.9 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 1.2 | 4.7 | GO:0042245 | RNA repair(GO:0042245) |
| 1.1 | 3.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 1.0 | 4.1 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 1.0 | 4.8 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 1.0 | 2.9 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.9 | 3.8 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.9 | 6.6 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.9 | 3.7 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.9 | 0.9 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.9 | 6.1 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.9 | 5.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.8 | 3.2 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.8 | 4.6 | GO:0050760 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.8 | 3.0 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.8 | 2.3 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.7 | 4.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.7 | 11.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.7 | 2.9 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.7 | 1.4 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.7 | 5.0 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.7 | 5.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.7 | 3.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.7 | 4.1 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.7 | 4.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.7 | 0.7 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.7 | 3.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.7 | 3.9 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.7 | 2.6 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.6 | 5.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.6 | 4.9 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.6 | 3.7 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.6 | 3.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.6 | 2.4 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.6 | 3.5 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.6 | 4.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.6 | 1.7 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.6 | 7.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.6 | 3.9 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.5 | 2.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.5 | 2.2 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.5 | 2.1 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.5 | 4.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.5 | 2.0 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.5 | 5.8 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.5 | 2.3 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.5 | 2.8 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) regulation of establishment of T cell polarity(GO:1903903) |
| 0.5 | 0.9 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.5 | 1.8 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.5 | 1.4 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.5 | 0.9 | GO:2000282 | regulation of cellular amino acid biosynthetic process(GO:2000282) |
| 0.4 | 1.3 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.4 | 1.3 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.4 | 1.3 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.4 | 3.8 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.4 | 5.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.4 | 3.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 2.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.4 | 2.5 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.4 | 2.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.4 | 2.9 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.4 | 1.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.4 | 0.4 | GO:0071415 | cellular response to purine-containing compound(GO:0071415) |
| 0.4 | 2.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.4 | 2.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 1.2 | GO:0021837 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.4 | 1.2 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.4 | 7.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.4 | 8.1 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.4 | 1.9 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.4 | 5.0 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.4 | 2.3 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.4 | 1.5 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.4 | 3.6 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.4 | 0.4 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) |
| 0.4 | 1.4 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.4 | 1.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.3 | 2.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.3 | 7.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.3 | 3.7 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.3 | 1.3 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.3 | 1.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.3 | 2.6 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.3 | 1.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 1.0 | GO:1903977 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) positive regulation of glial cell migration(GO:1903977) |
| 0.3 | 3.2 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.3 | 1.3 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.3 | 2.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.3 | 0.9 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.3 | 1.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.3 | 0.9 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.3 | 5.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 23.9 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.3 | 0.9 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.3 | 0.9 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.3 | 2.9 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 0.8 | GO:0002086 | diaphragm contraction(GO:0002086) vacuolar sequestering(GO:0043181) |
| 0.3 | 3.0 | GO:0015824 | proline transport(GO:0015824) |
| 0.3 | 2.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.3 | 6.9 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.3 | 0.8 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.3 | 1.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.3 | 1.3 | GO:0000101 | sulfur amino acid transport(GO:0000101) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 2.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.3 | 0.5 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.3 | 2.9 | GO:0015705 | iodide transport(GO:0015705) |
| 0.3 | 1.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.3 | 9.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.3 | 0.8 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.3 | 1.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.2 | 4.0 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.2 | 0.7 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 0.7 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 1.7 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 5.2 | GO:0030815 | negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cAMP metabolic process(GO:0030815) |
| 0.2 | 1.0 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.2 | 1.0 | GO:0090116 | DNA methylation on cytosine within a CG sequence(GO:0010424) C-5 methylation of cytosine(GO:0090116) |
| 0.2 | 1.7 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.2 | 1.7 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 2.4 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.2 | 5.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 1.0 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 0.7 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.2 | 2.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.2 | 3.5 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.2 | 1.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 0.7 | GO:1903400 | mitochondrial ornithine transport(GO:0000066) L-ornithine transmembrane transport(GO:1903352) L-arginine transmembrane transport(GO:1903400) L-lysine transmembrane transport(GO:1903401) arginine transmembrane transport(GO:1903826) |
| 0.2 | 3.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 | 15.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 0.9 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 5.4 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.2 | 1.6 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.2 | 5.6 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.2 | 1.6 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.2 | 0.7 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.2 | 1.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.2 | 0.9 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.2 | 0.9 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 0.9 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.2 | 0.4 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.2 | 0.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.2 | 1.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.2 | 0.4 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.2 | 2.8 | GO:0030432 | peristalsis(GO:0030432) |
| 0.2 | 1.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.2 | 8.6 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.2 | 2.7 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.2 | 1.9 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.2 | 3.5 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 2.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 2.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 1.9 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.2 | 4.1 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.2 | 0.9 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.2 | 1.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 1.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 0.5 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 1.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 0.7 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.2 | 1.6 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.2 | 3.5 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.2 | 0.9 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 | 0.5 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 | 0.8 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.2 | 0.7 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.2 | 4.7 | GO:0072606 | interleukin-8 secretion(GO:0072606) |
| 0.2 | 0.5 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.2 | 0.8 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 1.0 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.2 | 1.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 2.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 3.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.2 | 0.5 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.2 | 0.9 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 | 1.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 2.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.2 | 1.2 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.2 | 0.8 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 6.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 0.3 | GO:0070142 | synaptic vesicle budding(GO:0070142) |
| 0.2 | 1.5 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.2 | 6.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 0.4 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.1 | 0.6 | GO:0009826 | unidimensional cell growth(GO:0009826) susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 2.1 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.4 | GO:0010925 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) protein secretion by platelet(GO:0070560) |
| 0.1 | 0.6 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.4 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.1 | 1.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 2.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.7 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.4 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.1 | 1.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.5 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.7 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 5.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.6 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.1 | 1.8 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 | 0.5 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.1 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.6 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 1.1 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 0.6 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 2.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 2.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.7 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.9 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 1.6 | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) |
| 0.1 | 0.8 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 5.4 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.1 | 0.9 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 2.4 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.1 | 2.5 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 3.9 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 0.4 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.1 | 0.9 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 0.5 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.8 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 1.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.3 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 1.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.9 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 4.8 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.7 | GO:1903527 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 1.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 1.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 2.1 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 0.3 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 1.4 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.1 | 1.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 3.9 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.1 | 2.0 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.1 | 2.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.1 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 | 1.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.4 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.3 | GO:0043091 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.1 | 1.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.5 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.3 | GO:0051350 | negative regulation of lyase activity(GO:0051350) |
| 0.1 | 2.0 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 1.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.9 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.1 | 2.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.2 | GO:1900238 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.1 | 0.8 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 | 2.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.2 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.1 | 2.3 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.1 | 11.1 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 1.5 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 1.0 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 1.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.9 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 5.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.0 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 0.6 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 2.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.6 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.5 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.4 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.4 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 1.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.4 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 1.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.0 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.1 | 2.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.5 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.4 | GO:1990166 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 4.8 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 1.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.5 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.1 | 0.7 | GO:0046479 | glycolipid catabolic process(GO:0019377) glycosylceramide catabolic process(GO:0046477) glycosphingolipid catabolic process(GO:0046479) |
| 0.1 | 0.4 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.3 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 0.9 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.5 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.8 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.3 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 2.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 2.3 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 0.6 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 3.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 1.2 | GO:0071545 | inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.1 | 0.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.2 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 5.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 2.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 1.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 2.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 2.4 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.1 | 0.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.3 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 0.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.3 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.2 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 2.1 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.1 | 0.2 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 2.2 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 1.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 1.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 1.1 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.7 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 1.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.0 | 0.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 1.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.0 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 1.7 | GO:0060749 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.0 | 0.1 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) neuroblast migration(GO:0097402) |
| 0.0 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 2.5 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.8 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 2.5 | GO:0007270 | neuron-neuron synaptic transmission(GO:0007270) |
| 0.0 | 0.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.7 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 1.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 1.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.9 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.3 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.4 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.4 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.7 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 1.9 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.7 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.1 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.0 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.3 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.5 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.0 | 0.5 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.1 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.5 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 11.7 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.2 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.4 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.2 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.0 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 1.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.9 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.9 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.6 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.3 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 1.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 2.2 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 2.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.1 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.6 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 0.8 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.3 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.5 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 1.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0018003 | peptidyl-lysine N6-acetylation(GO:0018003) |
| 0.0 | 0.0 | GO:2000322 | regulation of glucocorticoid receptor signaling pathway(GO:2000322) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.4 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.2 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 1.0 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 1.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.7 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.3 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.2 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.5 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.0 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.3 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.9 | 3.7 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.9 | 2.6 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.8 | 3.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.8 | 2.3 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.7 | 17.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.6 | 17.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.6 | 8.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.6 | 6.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.5 | 3.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.5 | 2.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.5 | 2.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.5 | 4.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.5 | 4.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 2.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.4 | 1.8 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.4 | 1.3 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.4 | 2.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.4 | 3.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 1.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.4 | 1.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.4 | 2.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 8.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 3.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 1.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 2.5 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 1.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 3.4 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 1.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.3 | 0.3 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.3 | 5.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 9.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.3 | 2.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 0.8 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.3 | 13.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.3 | 9.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 1.1 | GO:0044393 | microspike(GO:0044393) |
| 0.3 | 6.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 2.4 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.3 | 6.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 3.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 1.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.2 | 1.0 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.2 | 20.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 11.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 13.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.9 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 1.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 1.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 1.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 5.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 0.6 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 2.7 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 4.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 2.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 0.7 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.2 | 1.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 1.0 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.2 | 8.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 2.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 0.9 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 1.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 3.8 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 3.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 0.7 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 63.3 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.2 | 5.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 14.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 2.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 12.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.4 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 2.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 4.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 5.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.8 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.9 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 1.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 1.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.6 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 3.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 2.8 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 3.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.2 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 6.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.5 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.9 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 1.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 2.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 6.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 6.3 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 0.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.0 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 0.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.6 | GO:1904949 | ATPase complex(GO:1904949) |
| 0.1 | 3.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 3.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 5.8 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 3.5 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 2.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 16.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 0.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 1.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 4.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 3.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 3.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 3.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 8.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 7.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 4.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 3.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.0 | 0.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 3.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 2.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 2.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.0 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.7 | 5.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 1.7 | 18.5 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 1.6 | 4.8 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.6 | 4.7 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 1.1 | 4.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.0 | 11.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 1.0 | 25.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.0 | 5.0 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.9 | 2.7 | GO:0051424 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) corticotropin-releasing hormone binding(GO:0051424) |
| 0.9 | 2.7 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.9 | 2.6 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.9 | 2.6 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.8 | 3.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.8 | 2.4 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.7 | 2.9 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.7 | 5.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.7 | 2.8 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.7 | 4.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.7 | 2.7 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.7 | 4.0 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.6 | 5.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.6 | 19.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.6 | 3.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.6 | 4.1 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.6 | 3.5 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.6 | 3.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.6 | 2.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.6 | 1.7 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.5 | 2.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.5 | 3.7 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.5 | 4.8 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.5 | 4.7 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.5 | 1.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.5 | 1.8 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.4 | 4.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 3.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.4 | 2.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.4 | 1.3 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.4 | 2.8 | GO:0001601 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.4 | 2.8 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.4 | 1.6 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.4 | 21.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.4 | 1.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.4 | 4.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.4 | 2.9 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.4 | 10.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.4 | 1.8 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.4 | 1.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 2.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.3 | 1.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.3 | 1.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.3 | 0.9 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.3 | 3.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 4.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 3.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.3 | 0.9 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.3 | 5.9 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.3 | 2.0 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.3 | 1.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.3 | 1.4 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.3 | 1.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 1.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.3 | 1.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 2.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 1.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 1.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 2.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.2 | 1.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 0.7 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 3.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.2 | 7.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 1.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 0.7 | GO:0005289 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.2 | 0.7 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.2 | 0.9 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 1.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 5.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 1.5 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.2 | 0.8 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 0.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 0.4 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 1.4 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.2 | 0.8 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 0.6 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 2.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 13.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 2.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 1.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 5.7 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 0.8 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 1.3 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 2.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 6.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 1.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 3.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 4.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 5.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 1.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 1.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 2.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 1.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 0.9 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.2 | 0.8 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 2.6 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.2 | 1.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 1.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 0.6 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.2 | 2.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 1.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.4 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.1 | 0.3 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 3.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 2.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.8 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.5 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 2.3 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.1 | 0.9 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 1.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 5.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 2.3 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 1.0 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 4.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 5.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 11.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.8 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 3.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.1 | 1.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.3 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 1.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 3.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 3.4 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 1.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 1.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 8.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 1.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.6 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 1.9 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 1.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 1.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 4.6 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 0.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 4.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 3.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 5.1 | GO:0022839 | ion gated channel activity(GO:0022839) |
| 0.1 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 1.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.4 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 4.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.9 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.9 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 6.7 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 0.6 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.7 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.0 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 1.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 7.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 1.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.2 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 0.3 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 1.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 1.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.2 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 15.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 4.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 1.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 4.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 1.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 2.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 6.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.5 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 1.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 4.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.2 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.0 | 1.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.9 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.0 | 0.4 | GO:1990446 | U1 snRNA binding(GO:0030619) U1 snRNP binding(GO:1990446) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 2.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.0 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 16.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 17.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 1.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.2 | 3.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 10.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 7.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 4.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 1.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 5.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 5.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.2 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 17.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.8 | 26.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.5 | 25.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 13.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 7.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 7.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 4.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 7.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 3.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 4.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 4.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 4.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 4.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 6.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 2.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 8.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 3.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 2.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 4.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 2.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 8.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 5.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 4.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 6.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 6.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 1.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 7.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 2.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 2.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.9 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 1.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 2.3 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.0 | 2.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.5 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.0 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 2.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |