Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
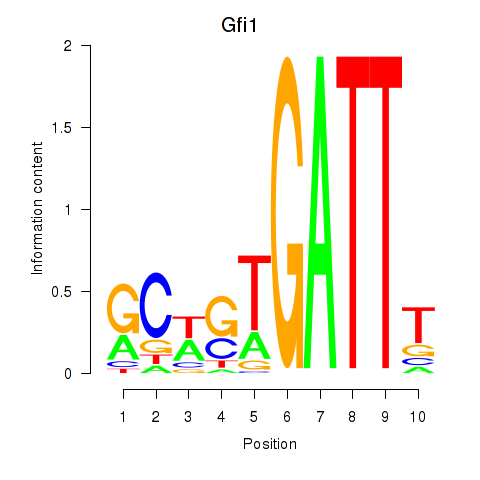
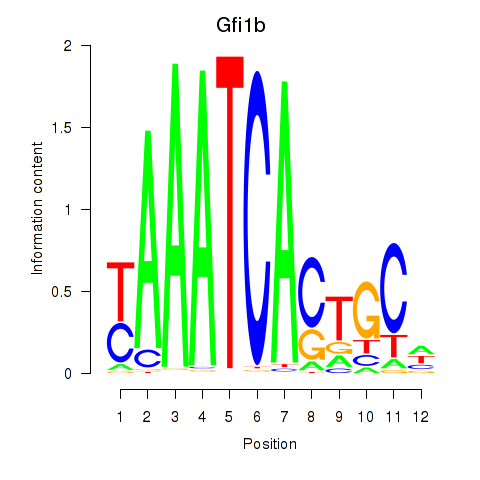
Results for Gfi1_Gfi1b
Z-value: 1.38
Transcription factors associated with Gfi1_Gfi1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gfi1
|
ENSMUSG00000029275.19 | Gfi1 |
|
Gfi1b
|
ENSMUSG00000026815.15 | Gfi1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gfi1b | mm39_v1_chr2_-_28511941_28511994 | -0.41 | 3.5e-04 | Click! |
| Gfi1 | mm39_v1_chr5_-_107873883_107873905 | -0.41 | 4.1e-04 | Click! |
Activity profile of Gfi1_Gfi1b motif
Sorted Z-values of Gfi1_Gfi1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gfi1_Gfi1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_43819688 | 9.67 |
ENSMUST00000048150.15
|
Cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr19_-_34231600 | 9.02 |
ENSMUST00000238147.2
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr16_+_92295009 | 8.84 |
ENSMUST00000023670.4
|
Clic6
|
chloride intracellular channel 6 |
| chr5_+_17779273 | 8.69 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr16_+_92089268 | 7.75 |
ENSMUST00000047383.10
|
Kcne2
|
potassium voltage-gated channel, Isk-related subfamily, gene 2 |
| chr3_+_54268523 | 7.62 |
ENSMUST00000117373.8
ENSMUST00000107985.10 ENSMUST00000073012.13 ENSMUST00000081564.13 |
Postn
|
periostin, osteoblast specific factor |
| chr3_+_92192724 | 6.55 |
ENSMUST00000193337.2
|
Sprr2a3
|
small proline-rich protein 2A3 |
| chr4_+_43406435 | 6.29 |
ENSMUST00000098106.9
ENSMUST00000139198.2 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr18_-_64688271 | 6.16 |
ENSMUST00000235459.2
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr1_-_169796709 | 5.97 |
ENSMUST00000027989.13
ENSMUST00000111353.4 |
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr7_-_99002204 | 5.58 |
ENSMUST00000208292.2
ENSMUST00000207989.2 ENSMUST00000208749.2 ENSMUST00000169437.9 ENSMUST00000208119.2 ENSMUST00000207849.2 |
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1 |
| chr17_+_35133435 | 5.49 |
ENSMUST00000007249.15
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr3_-_92366022 | 5.31 |
ENSMUST00000058142.4
|
Sprr3
|
small proline-rich protein 3 |
| chr11_+_101221431 | 5.30 |
ENSMUST00000103105.10
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr13_+_42454922 | 4.99 |
ENSMUST00000021796.9
|
Edn1
|
endothelin 1 |
| chr2_-_85349362 | 4.49 |
ENSMUST00000099923.2
|
Fads2b
|
fatty acid desaturase 2B |
| chr1_-_168259264 | 4.38 |
ENSMUST00000176790.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr16_-_97412169 | 4.24 |
ENSMUST00000232141.2
ENSMUST00000000395.8 |
Tmprss2
|
transmembrane protease, serine 2 |
| chr5_-_104261556 | 4.22 |
ENSMUST00000031249.8
|
Sparcl1
|
SPARC-like 1 |
| chr18_+_36475573 | 4.19 |
ENSMUST00000139727.3
ENSMUST00000237375.2 ENSMUST00000235403.2 ENSMUST00000236593.2 ENSMUST00000236374.2 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr19_-_32038838 | 4.18 |
ENSMUST00000096119.5
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr8_-_65146079 | 4.18 |
ENSMUST00000048967.9
|
Cpe
|
carboxypeptidase E |
| chr13_+_23755551 | 4.16 |
ENSMUST00000079251.8
|
H2bc8
|
H2B clustered histone 8 |
| chr9_-_67739607 | 3.97 |
ENSMUST00000054500.7
|
C2cd4a
|
C2 calcium-dependent domain containing 4A |
| chr11_+_101221895 | 3.97 |
ENSMUST00000017316.7
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr5_-_104261285 | 3.95 |
ENSMUST00000199947.2
|
Sparcl1
|
SPARC-like 1 |
| chr10_+_12966532 | 3.92 |
ENSMUST00000121646.8
ENSMUST00000121325.8 ENSMUST00000121766.8 |
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr3_+_92123106 | 3.84 |
ENSMUST00000074449.7
ENSMUST00000090871.3 |
Sprr2a1
Sprr2a2
|
small proline-rich protein 2A1 small proline-rich protein 2A2 |
| chr4_+_100633860 | 3.77 |
ENSMUST00000030257.15
ENSMUST00000097955.3 |
Cachd1
|
cache domain containing 1 |
| chr8_-_107007596 | 3.71 |
ENSMUST00000212896.2
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr5_+_37495801 | 3.64 |
ENSMUST00000056365.9
|
Evc2
|
EvC ciliary complex subunit 2 |
| chr1_-_168259070 | 3.59 |
ENSMUST00000064438.11
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr7_-_80055168 | 3.56 |
ENSMUST00000107362.10
ENSMUST00000135306.3 |
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr14_-_104081827 | 3.54 |
ENSMUST00000022718.11
|
Ednrb
|
endothelin receptor type B |
| chr5_+_14075281 | 3.50 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr7_-_89176294 | 3.41 |
ENSMUST00000207932.2
|
Prss23
|
protease, serine 23 |
| chr1_-_140111138 | 3.32 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr3_-_91990439 | 3.32 |
ENSMUST00000058150.8
|
Lor
|
loricrin |
| chr19_+_11724913 | 3.32 |
ENSMUST00000025585.4
|
Cblif
|
cobalamin binding intrinsic factor |
| chr11_-_99383938 | 3.32 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr8_-_13446769 | 3.29 |
ENSMUST00000033826.4
|
Atp4b
|
ATPase, H+/K+ exchanging, beta polypeptide |
| chr1_-_140111018 | 3.27 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr1_-_168259839 | 3.26 |
ENSMUST00000188912.7
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr7_-_81143631 | 3.23 |
ENSMUST00000082090.15
|
Ap3b2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr14_-_119336809 | 3.20 |
ENSMUST00000156203.8
|
Uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr5_-_44259010 | 3.20 |
ENSMUST00000087441.11
|
Prom1
|
prominin 1 |
| chr6_+_135339543 | 3.19 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr1_-_169575203 | 3.17 |
ENSMUST00000027991.12
ENSMUST00000111357.2 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr3_+_124114504 | 3.10 |
ENSMUST00000058994.6
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr16_+_44914397 | 3.10 |
ENSMUST00000061050.6
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr10_-_62067026 | 3.06 |
ENSMUST00000047883.11
|
Tspan15
|
tetraspanin 15 |
| chr9_+_45230370 | 3.05 |
ENSMUST00000034597.8
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr8_-_62044164 | 3.05 |
ENSMUST00000135439.2
ENSMUST00000121200.9 |
Palld
|
palladin, cytoskeletal associated protein |
| chr11_-_83959999 | 3.03 |
ENSMUST00000138208.2
|
Dusp14
|
dual specificity phosphatase 14 |
| chr6_-_119085508 | 2.98 |
ENSMUST00000188522.7
|
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr6_+_15185399 | 2.96 |
ENSMUST00000115474.8
ENSMUST00000115472.8 ENSMUST00000031545.14 |
Foxp2
|
forkhead box P2 |
| chr16_+_44913974 | 2.95 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr5_+_65357034 | 2.94 |
ENSMUST00000203653.3
ENSMUST00000041892.13 |
Wdr19
|
WD repeat domain 19 |
| chr1_-_168259465 | 2.93 |
ENSMUST00000176540.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr1_-_168259710 | 2.93 |
ENSMUST00000072863.6
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr9_+_66620959 | 2.85 |
ENSMUST00000071889.13
|
Car12
|
carbonic anhydrase 12 |
| chr18_-_43925932 | 2.84 |
ENSMUST00000237926.2
ENSMUST00000096570.4 |
Gm94
|
predicted gene 94 |
| chr9_+_66621001 | 2.81 |
ENSMUST00000085420.12
|
Car12
|
carbonic anhydrase 12 |
| chr2_+_143757193 | 2.77 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr7_+_121464254 | 2.77 |
ENSMUST00000033161.7
|
Scnn1b
|
sodium channel, nonvoltage-gated 1 beta |
| chr4_+_140737955 | 2.75 |
ENSMUST00000071977.9
|
Mfap2
|
microfibrillar-associated protein 2 |
| chr15_-_78739717 | 2.74 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr12_+_31488208 | 2.74 |
ENSMUST00000001254.6
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr7_-_59654849 | 2.74 |
ENSMUST00000059305.17
|
Snrpn
|
small nuclear ribonucleoprotein N |
| chr10_+_127595639 | 2.72 |
ENSMUST00000128247.2
|
Rdh16f1
|
RDH16 family member 1 |
| chr5_+_16139683 | 2.69 |
ENSMUST00000167946.9
|
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr7_-_144232586 | 2.63 |
ENSMUST00000131731.2
|
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr11_+_70431063 | 2.62 |
ENSMUST00000018429.12
ENSMUST00000108557.10 ENSMUST00000108556.2 |
Pld2
|
phospholipase D2 |
| chr15_+_10952418 | 2.61 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr16_-_91525863 | 2.58 |
ENSMUST00000073466.13
|
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr9_-_99592116 | 2.57 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr8_+_46338498 | 2.56 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr7_-_81143412 | 2.55 |
ENSMUST00000238438.2
ENSMUST00000238711.2 |
Ap3b2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr1_+_133278248 | 2.55 |
ENSMUST00000094556.3
|
Ren1
|
renin 1 structural |
| chr4_+_128999325 | 2.53 |
ENSMUST00000106064.10
ENSMUST00000030575.15 ENSMUST00000030577.11 |
Tmem54
|
transmembrane protein 54 |
| chr8_+_107662352 | 2.51 |
ENSMUST00000212524.2
ENSMUST00000047425.5 |
Sntb2
|
syntrophin, basic 2 |
| chr7_-_129867967 | 2.50 |
ENSMUST00000117872.8
ENSMUST00000120187.9 |
Fgfr2
|
fibroblast growth factor receptor 2 |
| chr2_-_120144622 | 2.49 |
ENSMUST00000054651.8
|
Pla2g4f
|
phospholipase A2, group IVF |
| chr3_+_121085471 | 2.48 |
ENSMUST00000199554.2
|
Alg14
|
asparagine-linked glycosylation 14 |
| chr8_-_118400418 | 2.47 |
ENSMUST00000173522.8
ENSMUST00000174450.2 |
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr6_-_52185674 | 2.43 |
ENSMUST00000062829.9
|
Hoxa6
|
homeobox A6 |
| chr3_+_59832635 | 2.43 |
ENSMUST00000049476.3
|
Aadacl2fm1
|
AADACL2 family member 1 |
| chr4_+_116453927 | 2.40 |
ENSMUST00000051869.8
|
Ccdc17
|
coiled-coil domain containing 17 |
| chr4_+_9269285 | 2.39 |
ENSMUST00000038841.14
|
Clvs1
|
clavesin 1 |
| chr10_+_34359395 | 2.38 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr7_-_59654797 | 2.35 |
ENSMUST00000194059.2
|
Snurf
|
SNRPN upstream reading frame |
| chr10_+_34359513 | 2.34 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chrX_-_74918709 | 2.34 |
ENSMUST00000114059.10
|
Pls3
|
plastin 3 (T-isoform) |
| chr8_+_46338557 | 2.33 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr1_+_185064339 | 2.33 |
ENSMUST00000027916.13
ENSMUST00000210277.2 ENSMUST00000151769.2 ENSMUST00000110965.2 |
Bpnt1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr2_+_49956441 | 2.33 |
ENSMUST00000112712.10
ENSMUST00000128451.8 ENSMUST00000053208.14 |
Lypd6
|
LY6/PLAUR domain containing 6 |
| chr10_+_125802084 | 2.31 |
ENSMUST00000074807.8
|
Lrig3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr3_+_59914164 | 2.30 |
ENSMUST00000169794.2
|
Aadacl2
|
arylacetamide deacetylase like 2 |
| chr3_+_92272486 | 2.30 |
ENSMUST00000050397.2
|
Sprr2f
|
small proline-rich protein 2F |
| chr3_-_121325887 | 2.29 |
ENSMUST00000039197.9
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr19_-_20704896 | 2.28 |
ENSMUST00000025656.4
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr3_+_68479578 | 2.24 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr6_+_17306334 | 2.23 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr6_+_29853745 | 2.19 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr9_+_72600721 | 2.19 |
ENSMUST00000238315.2
|
Nedd4
|
neural precursor cell expressed, developmentally down-regulated 4 |
| chr6_-_92458324 | 2.18 |
ENSMUST00000113445.2
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr12_-_111344139 | 2.18 |
ENSMUST00000041965.5
|
Cdc42bpb
|
CDC42 binding protein kinase beta |
| chr6_+_34575435 | 2.17 |
ENSMUST00000079391.10
ENSMUST00000142512.8 ENSMUST00000115027.8 ENSMUST00000115026.8 |
Cald1
|
caldesmon 1 |
| chr8_-_37419567 | 2.17 |
ENSMUST00000163663.3
|
Dlc1
|
deleted in liver cancer 1 |
| chr9_-_31824758 | 2.12 |
ENSMUST00000116615.5
|
Barx2
|
BarH-like homeobox 2 |
| chr1_+_11063678 | 2.11 |
ENSMUST00000027056.12
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr3_-_49711706 | 2.11 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr3_+_121085373 | 2.10 |
ENSMUST00000039442.12
|
Alg14
|
asparagine-linked glycosylation 14 |
| chr16_-_97306125 | 2.09 |
ENSMUST00000049721.9
ENSMUST00000231999.2 |
Fam3b
|
family with sequence similarity 3, member B |
| chr14_-_45767421 | 2.09 |
ENSMUST00000150660.3
|
Fermt2
|
fermitin family member 2 |
| chr4_-_135300934 | 2.08 |
ENSMUST00000105855.2
|
Grhl3
|
grainyhead like transcription factor 3 |
| chr11_-_68762664 | 2.05 |
ENSMUST00000101017.9
|
Ndel1
|
nudE neurodevelopment protein 1 like 1 |
| chr1_-_45542442 | 2.05 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr7_+_4467730 | 2.05 |
ENSMUST00000086372.8
ENSMUST00000169820.8 ENSMUST00000163893.8 ENSMUST00000170635.2 |
Eps8l1
|
EPS8-like 1 |
| chr3_+_121085500 | 2.04 |
ENSMUST00000198341.2
|
Alg14
|
asparagine-linked glycosylation 14 |
| chr7_-_89167079 | 2.02 |
ENSMUST00000207538.2
|
Prss23
|
protease, serine 23 |
| chr17_-_46343291 | 1.99 |
ENSMUST00000071648.12
ENSMUST00000142351.9 |
Vegfa
|
vascular endothelial growth factor A |
| chrX_-_46981379 | 1.99 |
ENSMUST00000077569.11
ENSMUST00000101616.9 ENSMUST00000088973.11 |
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr5_-_124003553 | 1.99 |
ENSMUST00000057145.7
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chr16_-_45773929 | 1.97 |
ENSMUST00000134802.8
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr2_+_27776428 | 1.97 |
ENSMUST00000028280.14
|
Col5a1
|
collagen, type V, alpha 1 |
| chr5_+_93073259 | 1.95 |
ENSMUST00000172706.2
|
Shroom3
|
shroom family member 3 |
| chr9_+_50466127 | 1.92 |
ENSMUST00000213916.2
|
Il18
|
interleukin 18 |
| chr5_+_92831150 | 1.89 |
ENSMUST00000113055.9
|
Shroom3
|
shroom family member 3 |
| chr9_-_79885063 | 1.88 |
ENSMUST00000093811.11
|
Filip1
|
filamin A interacting protein 1 |
| chr9_-_79884920 | 1.88 |
ENSMUST00000239133.2
|
Filip1
|
filamin A interacting protein 1 |
| chr3_-_49711765 | 1.87 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chrX_+_138464065 | 1.87 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr19_-_36714053 | 1.86 |
ENSMUST00000087321.4
|
Ppp1r3c
|
protein phosphatase 1, regulatory subunit 3C |
| chr5_-_103777145 | 1.86 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr8_-_62576140 | 1.85 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr11_-_109188947 | 1.84 |
ENSMUST00000020920.10
|
Rgs9
|
regulator of G-protein signaling 9 |
| chrX_+_163156359 | 1.80 |
ENSMUST00000033751.8
|
Vegfd
|
vascular endothelial growth factor D |
| chr3_+_32871669 | 1.80 |
ENSMUST00000072312.12
ENSMUST00000108228.8 |
Usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr10_+_94411119 | 1.76 |
ENSMUST00000121471.8
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr2_-_80411724 | 1.76 |
ENSMUST00000111760.3
|
Nckap1
|
NCK-associated protein 1 |
| chr19_-_9065309 | 1.76 |
ENSMUST00000025554.3
|
Scgb1a1
|
secretoglobin, family 1A, member 1 (uteroglobin) |
| chr1_+_152275575 | 1.76 |
ENSMUST00000044311.9
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr18_-_20247666 | 1.74 |
ENSMUST00000224432.2
|
Dsc1
|
desmocollin 1 |
| chr17_+_12338161 | 1.73 |
ENSMUST00000024594.9
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chrX_-_142610371 | 1.72 |
ENSMUST00000087316.6
|
Capn6
|
calpain 6 |
| chr2_-_57942844 | 1.72 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein |
| chr5_+_32768515 | 1.71 |
ENSMUST00000202543.4
ENSMUST00000072311.13 |
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr17_+_43978377 | 1.71 |
ENSMUST00000233627.2
ENSMUST00000233437.2 |
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr6_-_113911640 | 1.70 |
ENSMUST00000101044.9
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr16_+_84571011 | 1.70 |
ENSMUST00000114195.8
|
Jam2
|
junction adhesion molecule 2 |
| chr4_-_129155185 | 1.69 |
ENSMUST00000145261.8
|
C77080
|
expressed sequence C77080 |
| chr1_-_158183894 | 1.68 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr9_+_70114623 | 1.67 |
ENSMUST00000034745.9
|
Myo1e
|
myosin IE |
| chr2_+_20524587 | 1.66 |
ENSMUST00000114604.9
ENSMUST00000066509.10 |
Etl4
|
enhancer trap locus 4 |
| chr9_-_67336085 | 1.66 |
ENSMUST00000213584.2
|
Tln2
|
talin 2 |
| chr12_-_111344038 | 1.65 |
ENSMUST00000222196.2
|
Cdc42bpb
|
CDC42 binding protein kinase beta |
| chr18_+_65715156 | 1.64 |
ENSMUST00000237553.2
ENSMUST00000237712.2 ENSMUST00000182684.8 |
Zfp532
|
zinc finger protein 532 |
| chrX_-_162810959 | 1.63 |
ENSMUST00000033739.5
|
Car5b
|
carbonic anhydrase 5b, mitochondrial |
| chr17_-_24662055 | 1.61 |
ENSMUST00000119932.8
ENSMUST00000088506.12 |
Dnase1l2
|
deoxyribonuclease 1-like 2 |
| chr10_+_127595590 | 1.61 |
ENSMUST00000073639.6
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr7_+_49624612 | 1.60 |
ENSMUST00000151721.8
ENSMUST00000081872.13 |
Nell1
|
NEL-like 1 |
| chr4_-_57300361 | 1.58 |
ENSMUST00000153926.8
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr1_+_17672117 | 1.57 |
ENSMUST00000088476.4
|
Pi15
|
peptidase inhibitor 15 |
| chr5_-_66776095 | 1.56 |
ENSMUST00000162366.8
ENSMUST00000162994.8 ENSMUST00000159512.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chrX_-_46981273 | 1.56 |
ENSMUST00000153548.9
ENSMUST00000141084.3 |
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr16_-_45664591 | 1.56 |
ENSMUST00000076333.12
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr6_+_30568366 | 1.56 |
ENSMUST00000049251.6
|
Cpa4
|
carboxypeptidase A4 |
| chr10_-_127177729 | 1.55 |
ENSMUST00000026474.5
ENSMUST00000219671.2 |
Gli1
|
GLI-Kruppel family member GLI1 |
| chr2_-_64806106 | 1.55 |
ENSMUST00000156765.2
|
Grb14
|
growth factor receptor bound protein 14 |
| chr14_-_45767232 | 1.55 |
ENSMUST00000149723.2
|
Fermt2
|
fermitin family member 2 |
| chr10_-_12745109 | 1.55 |
ENSMUST00000218635.2
|
Utrn
|
utrophin |
| chr18_+_5593566 | 1.54 |
ENSMUST00000160910.2
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr13_-_78344492 | 1.54 |
ENSMUST00000125176.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr16_+_91526169 | 1.54 |
ENSMUST00000114001.8
ENSMUST00000113999.8 ENSMUST00000064797.12 ENSMUST00000114002.9 ENSMUST00000095909.10 ENSMUST00000056482.14 ENSMUST00000113996.8 |
Itsn1
|
intersectin 1 (SH3 domain protein 1A) |
| chr2_-_144392696 | 1.54 |
ENSMUST00000028915.6
|
Rbbp9
|
retinoblastoma binding protein 9, serine hydrolase |
| chr16_+_25105560 | 1.54 |
ENSMUST00000056087.4
|
Tprg
|
transformation related protein 63 regulated |
| chr17_+_43978280 | 1.52 |
ENSMUST00000170988.2
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr19_+_38825005 | 1.52 |
ENSMUST00000037302.6
|
Tbc1d12
|
TBC1D12: TBC1 domain family, member 12 |
| chr15_-_55770312 | 1.52 |
ENSMUST00000039769.13
|
Sntb1
|
syntrophin, basic 1 |
| chr3_+_159545309 | 1.51 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr15_+_78915071 | 1.50 |
ENSMUST00000006544.9
ENSMUST00000171999.9 |
Gcat
Gcat
|
glycine C-acetyltransferase (2-amino-3-ketobutyrate-coenzyme A ligase) glycine C-acetyltransferase (2-amino-3-ketobutyrate-coenzyme A ligase) |
| chr3_-_27950491 | 1.50 |
ENSMUST00000058077.4
|
Tmem212
|
transmembrane protein 212 |
| chr17_+_7437500 | 1.49 |
ENSMUST00000024575.8
|
Rps6ka2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr15_+_100202061 | 1.49 |
ENSMUST00000229574.2
ENSMUST00000229217.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr19_-_10857734 | 1.49 |
ENSMUST00000133303.8
|
Tmem109
|
transmembrane protein 109 |
| chr11_-_31621727 | 1.45 |
ENSMUST00000109415.2
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr15_-_13173736 | 1.44 |
ENSMUST00000036439.6
|
Cdh6
|
cadherin 6 |
| chrX_+_99811325 | 1.44 |
ENSMUST00000000901.13
ENSMUST00000113736.9 ENSMUST00000087984.11 |
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chrX_+_141009756 | 1.44 |
ENSMUST00000112916.9
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr19_-_34855278 | 1.43 |
ENSMUST00000112460.3
|
Pank1
|
pantothenate kinase 1 |
| chr17_-_46342739 | 1.42 |
ENSMUST00000024747.14
|
Vegfa
|
vascular endothelial growth factor A |
| chr9_+_21838767 | 1.42 |
ENSMUST00000006403.7
ENSMUST00000170304.9 ENSMUST00000216710.2 |
Ccdc159
|
coiled-coil domain containing 159 |
| chr3_-_89009153 | 1.41 |
ENSMUST00000199668.3
ENSMUST00000196709.5 |
Fdps
|
farnesyl diphosphate synthetase |
| chr18_+_37533899 | 1.41 |
ENSMUST00000057228.2
|
Pcdhb9
|
protocadherin beta 9 |
| chr3_-_57755500 | 1.41 |
ENSMUST00000066882.10
|
Pfn2
|
profilin 2 |
| chr6_+_127864543 | 1.41 |
ENSMUST00000032501.6
|
Tspan11
|
tetraspanin 11 |
| chr11_+_66915969 | 1.40 |
ENSMUST00000079077.12
ENSMUST00000061786.6 |
Tmem220
|
transmembrane protein 220 |
| chr3_+_102641822 | 1.40 |
ENSMUST00000029451.12
|
Tspan2
|
tetraspanin 2 |
| chr15_-_26895660 | 1.39 |
ENSMUST00000059204.11
|
Fbxl7
|
F-box and leucine-rich repeat protein 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 1.9 | 9.7 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 1.9 | 7.6 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.8 | 5.5 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 1.7 | 5.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 1.7 | 5.0 | GO:0030070 | insulin processing(GO:0030070) |
| 1.1 | 3.4 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 1.1 | 3.3 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 1.1 | 8.7 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 1.1 | 3.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 1.0 | 9.3 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 1.0 | 4.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.0 | 9.0 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 1.0 | 2.9 | GO:0060032 | notochord regression(GO:0060032) |
| 0.9 | 3.6 | GO:0042335 | cuticle development(GO:0042335) |
| 0.9 | 3.6 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.9 | 3.5 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.8 | 2.5 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) |
| 0.8 | 2.4 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 0.8 | 6.2 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.8 | 2.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.7 | 2.9 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.7 | 6.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.7 | 2.7 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.6 | 3.2 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.6 | 3.7 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.6 | 2.3 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.6 | 4.6 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.5 | 2.7 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.5 | 2.6 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.5 | 1.6 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.5 | 5.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.5 | 5.6 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.5 | 1.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.5 | 3.0 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.5 | 3.4 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.5 | 1.9 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.5 | 2.8 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.5 | 2.7 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.4 | 1.8 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 0.4 | 17.0 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.4 | 0.9 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.4 | 6.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.4 | 1.8 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.4 | 2.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.4 | 1.7 | GO:0048840 | otolith development(GO:0048840) |
| 0.4 | 1.2 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.4 | 1.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.4 | 1.2 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.4 | 4.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.4 | 2.2 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.4 | 2.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.4 | 1.1 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.3 | 1.4 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.3 | 1.4 | GO:0032379 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.3 | 2.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.3 | 1.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.3 | 1.3 | GO:0035565 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.3 | 3.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 0.9 | GO:0061193 | taste bud development(GO:0061193) |
| 0.3 | 1.8 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.3 | 0.9 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.3 | 11.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.3 | 1.8 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.3 | 1.8 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.3 | 0.6 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.3 | 0.3 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.3 | 1.1 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.3 | 1.4 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.3 | 6.5 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.3 | 5.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.3 | 0.8 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.3 | 1.0 | GO:0034971 | histone H3-R17 methylation(GO:0034971) bile acid signaling pathway(GO:0038183) |
| 0.2 | 2.7 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.2 | 2.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 2.9 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.2 | 1.2 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.2 | 2.1 | GO:1904729 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal lipid absorption(GO:1904729) |
| 0.2 | 0.9 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.2 | 1.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 1.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.2 | 0.7 | GO:2000184 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.2 | 1.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.2 | 1.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 4.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 1.5 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.2 | 2.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 0.9 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.2 | 0.8 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.2 | 1.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 1.0 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.2 | 0.6 | GO:1904268 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.2 | 2.1 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.2 | 0.6 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.2 | 3.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 0.6 | GO:1904172 | positive regulation of bleb assembly(GO:1904172) |
| 0.2 | 0.7 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 1.7 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.2 | 0.7 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.2 | 1.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 1.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 9.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 0.9 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.2 | 1.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 0.5 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.2 | 1.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.2 | 0.7 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.2 | 1.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 0.5 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 | 0.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.2 | 0.8 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.2 | 0.6 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.2 | 1.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.4 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 1.5 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.1 | 0.7 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 10.3 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.1 | 10.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.3 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 | 0.6 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.1 | 1.7 | GO:0042637 | catagen(GO:0042637) |
| 0.1 | 0.8 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.1 | 1.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.6 | GO:1903943 | estrous cycle(GO:0044849) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 1.5 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 4.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 5.7 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.1 | 0.5 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 2.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 1.5 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.7 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 2.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 3.3 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.1 | 2.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.6 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 3.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 2.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 3.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.8 | GO:1903758 | regulation of postsynaptic density protein 95 clustering(GO:1902897) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 2.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 3.6 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.7 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 1.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 1.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 1.1 | GO:0035931 | mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.1 | 1.0 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.1 | 0.7 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.6 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.5 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 1.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.3 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.3 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 1.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.2 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.1 | 0.6 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 2.5 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.1 | 2.0 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.1 | 0.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 1.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 1.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 1.4 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 1.1 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 2.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.4 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.1 | 1.1 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.5 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.1 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 | 3.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 3.2 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 1.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.9 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 0.4 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.3 | GO:0055048 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 1.0 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 1.2 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 0.3 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) amino acid homeostasis(GO:0080144) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.1 | 1.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.3 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.2 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.1 | 0.4 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 2.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.1 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.1 | 0.1 | GO:0003195 | tricuspid valve formation(GO:0003195) |
| 0.1 | 2.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 2.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.5 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 1.7 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.2 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.6 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.1 | 0.9 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.2 | GO:0070781 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.0 | 6.0 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 3.5 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 2.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.9 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:1900275 | actin filament branching(GO:0090135) negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 1.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 2.2 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 1.8 | GO:0006760 | folic acid-containing compound metabolic process(GO:0006760) |
| 0.0 | 2.8 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 1.8 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 3.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0060873 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) frontal suture morphogenesis(GO:0060364) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.0 | 0.6 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.7 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 2.2 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.2 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.3 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 1.0 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 2.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 2.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 4.2 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 0.6 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 1.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 2.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.4 | GO:1902572 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.5 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 2.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 | 1.4 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.4 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.5 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 1.7 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 2.5 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.2 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.2 | GO:0006664 | glycolipid metabolic process(GO:0006664) |
| 0.0 | 0.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 2.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.4 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 1.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 2.6 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 2.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 1.7 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.5 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 0.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 3.1 | GO:0051604 | protein maturation(GO:0051604) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.2 | 3.5 | GO:0090537 | CERF complex(GO:0090537) |
| 1.0 | 9.0 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.0 | 3.0 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 0.7 | 5.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.5 | 9.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.5 | 3.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.4 | 21.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 5.8 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.3 | 3.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 1.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.3 | 3.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 3.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 3.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.3 | 0.9 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 1.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.3 | 0.8 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.3 | 3.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 1.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 4.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 0.7 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.2 | 2.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 1.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 4.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 0.8 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.2 | 2.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 3.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 6.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.6 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 2.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 1.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 1.8 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 1.9 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 2.0 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.2 | 2.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 0.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.1 | GO:0030897 | HOPS complex(GO:0030897) FHF complex(GO:0070695) |
| 0.1 | 0.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 11.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.5 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 1.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.4 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 2.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 5.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 12.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 8.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 2.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.4 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 5.6 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.1 | 0.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 0.6 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 1.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 16.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.2 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.1 | 1.7 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.3 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 5.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 4.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.2 | GO:0005712 | chiasma(GO:0005712) late recombination nodule(GO:0005715) |
| 0.1 | 0.5 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.5 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 2.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 4.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 7.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 5.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 10.9 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 1.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.1 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 4.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 4.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.0 | 1.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 3.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 1.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 2.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 7.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 3.0 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.5 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 4.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.0 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 10.9 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.2 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.5 | GO:0005685 | U1 snRNP(GO:0005685) U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 1.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 7.1 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0030663 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 3.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.2 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 3.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 32.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 2.9 | 8.8 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 2.0 | 6.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 1.2 | 5.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.2 | 3.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 1.1 | 3.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.1 | 3.3 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 1.1 | 3.2 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.9 | 6.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.9 | 2.7 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.9 | 7.7 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.8 | 5.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.7 | 12.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.7 | 2.7 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.6 | 6.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.6 | 2.3 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.6 | 3.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.5 | 2.7 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.5 | 4.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.5 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.5 | 2.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.4 | 2.6 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.4 | 3.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.4 | 2.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.4 | 2.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.4 | 7.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 1.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.4 | 1.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.4 | 2.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.4 | 3.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.4 | 2.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.4 | 1.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 4.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 1.4 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.3 | 1.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.3 | 2.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 1.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.3 | 3.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.3 | 2.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 0.9 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 2.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 3.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 3.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 1.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.3 | 0.3 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.3 | 2.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 1.8 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.3 | 1.0 | GO:0051381 | histamine binding(GO:0051381) |
| 0.2 | 0.7 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 2.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 1.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 2.8 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 2.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.2 | 1.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 1.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.2 | 3.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 6.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 0.7 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.2 | 1.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.2 | 3.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 1.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.2 | 1.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 2.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 1.1 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.2 | 0.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 2.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 1.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 14.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 0.6 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 0.9 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.9 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 2.0 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.8 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 2.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.4 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.1 | 1.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.6 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 4.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.3 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.9 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.8 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 2.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.8 | GO:0002135 | CTP binding(GO:0002135) sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 7.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.3 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 3.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.8 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 1.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 4.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 0.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 2.7 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.1 | 2.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.9 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.1 | 1.0 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 3.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 5.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 1.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 2.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 3.3 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 1.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 1.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 4.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 2.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.5 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 2.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 2.2 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 1.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 10.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.8 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 5.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 7.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 1.6 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.1 | 2.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 2.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.2 | GO:0018271 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.1 | 2.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 2.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 2.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.0 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 2.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 7.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.8 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 2.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 16.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 4.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 1.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.1 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 0.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 3.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 1.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 9.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 8.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 2.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 10.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 3.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 16.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 21.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 7.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 3.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 9.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 18.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 1.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 0.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 3.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 2.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 7.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 2.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 2.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 2.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 4.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 3.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 10.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 3.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 7.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 11.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 3.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 3.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 6.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 5.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 11.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 8.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 2.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 11.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 3.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 2.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 4.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 3.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 3.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 1.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.5 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 2.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 2.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 3.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 0.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.7 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 0.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.1 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 2.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 2.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 4.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.3 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.3 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 1.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.2 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 1.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.8 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.4 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |