Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Gli1
Z-value: 1.07
Transcription factors associated with Gli1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli1
|
ENSMUSG00000025407.8 | Gli1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gli1 | mm39_v1_chr10_-_127177729_127177843 | 0.24 | 4.7e-02 | Click! |
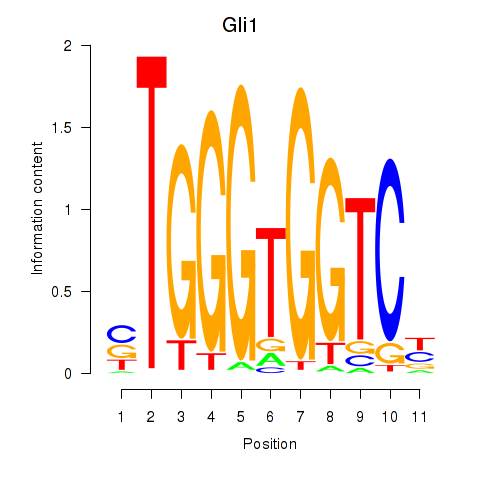
Activity profile of Gli1 motif
Sorted Z-values of Gli1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gli1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_34457868 | 10.84 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr11_+_72889889 | 6.08 |
ENSMUST00000021141.14
|
P2rx1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr7_+_24069680 | 6.00 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr11_+_115790768 | 4.96 |
ENSMUST00000152171.8
|
Smim5
|
small integral membrane protein 5 |
| chr12_-_113561594 | 4.58 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr6_+_113508636 | 4.46 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr7_-_24459736 | 4.44 |
ENSMUST00000063956.7
|
Cd177
|
CD177 antigen |
| chr12_-_113823290 | 4.31 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr4_-_133601990 | 4.26 |
ENSMUST00000168974.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr3_+_90511068 | 4.25 |
ENSMUST00000001046.7
|
S100a4
|
S100 calcium binding protein A4 |
| chrX_+_158086253 | 4.09 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chrX_-_166165743 | 4.05 |
ENSMUST00000026839.5
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr9_+_106083988 | 4.01 |
ENSMUST00000188650.2
|
Twf2
|
twinfilin actin binding protein 2 |
| chr2_+_117942357 | 3.98 |
ENSMUST00000039559.9
|
Thbs1
|
thrombospondin 1 |
| chr6_-_148846247 | 3.85 |
ENSMUST00000111562.8
ENSMUST00000081956.12 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr11_+_115353290 | 3.78 |
ENSMUST00000106532.4
ENSMUST00000092445.12 ENSMUST00000153466.2 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr3_+_92195873 | 3.70 |
ENSMUST00000090872.7
|
Sprr2a3
|
small proline-rich protein 2A3 |
| chr12_-_114117264 | 3.66 |
ENSMUST00000103461.5
|
Ighv7-3
|
immunoglobulin heavy variable 7-3 |
| chr17_+_37356854 | 3.63 |
ENSMUST00000025338.16
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr12_-_113294138 | 3.51 |
ENSMUST00000194304.6
ENSMUST00000103420.3 |
Ighg1
|
immunoglobulin heavy constant gamma 1 (G1m marker) |
| chr9_-_106762818 | 3.27 |
ENSMUST00000185707.2
|
Rbm15b
|
RNA binding motif protein 15B |
| chr12_-_113625906 | 3.27 |
ENSMUST00000103448.3
|
Ighv5-9
|
immunoglobulin heavy variable 5-9 |
| chr6_+_48603698 | 3.22 |
ENSMUST00000095938.10
|
AI854703
|
expressed sequence AI854703 |
| chr1_-_64160557 | 3.05 |
ENSMUST00000055001.10
ENSMUST00000114086.8 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr14_-_54646917 | 3.04 |
ENSMUST00000000984.9
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr5_-_115439016 | 3.03 |
ENSMUST00000009157.4
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr18_-_70274639 | 3.02 |
ENSMUST00000121693.8
|
Rab27b
|
RAB27B, member RAS oncogene family |
| chr11_+_76900091 | 3.00 |
ENSMUST00000129572.3
|
Slc6a4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
| chr11_-_102255999 | 2.89 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr7_+_79836581 | 2.88 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr11_-_34724458 | 2.88 |
ENSMUST00000093191.3
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr7_-_19504446 | 2.88 |
ENSMUST00000003061.14
|
Bcam
|
basal cell adhesion molecule |
| chr10_+_79722081 | 2.88 |
ENSMUST00000046091.7
|
Elane
|
elastase, neutrophil expressed |
| chr5_-_115438971 | 2.85 |
ENSMUST00000112090.2
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr5_-_110987604 | 2.84 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr3_+_95836558 | 2.82 |
ENSMUST00000165307.8
ENSMUST00000015893.13 ENSMUST00000169426.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr7_+_28682253 | 2.81 |
ENSMUST00000085835.8
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr15_-_100567377 | 2.75 |
ENSMUST00000182814.8
ENSMUST00000238935.2 ENSMUST00000182068.8 ENSMUST00000182574.2 ENSMUST00000182775.8 |
Bin2
|
bridging integrator 2 |
| chr3_+_95836637 | 2.67 |
ENSMUST00000171368.8
ENSMUST00000168106.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr6_-_41613322 | 2.67 |
ENSMUST00000031902.7
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr1_-_106641940 | 2.64 |
ENSMUST00000112751.2
|
Bcl2
|
B cell leukemia/lymphoma 2 |
| chr4_+_62398262 | 2.64 |
ENSMUST00000030088.12
ENSMUST00000107449.4 |
Bspry
|
B-box and SPRY domain containing |
| chr4_+_12089373 | 2.61 |
ENSMUST00000095143.9
ENSMUST00000063839.6 |
Rbm12b2
|
RNA binding motif protein 12 B2 |
| chr11_-_83353787 | 2.58 |
ENSMUST00000021020.13
ENSMUST00000119346.2 ENSMUST00000103209.10 ENSMUST00000108137.9 |
Mmp28
|
matrix metallopeptidase 28 (epilysin) |
| chr13_+_44882998 | 2.58 |
ENSMUST00000174068.8
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr2_-_91762033 | 2.51 |
ENSMUST00000111309.8
ENSMUST00000090602.6 |
Mdk
|
midkine |
| chr17_+_37356872 | 2.39 |
ENSMUST00000174456.8
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr12_+_17778198 | 2.36 |
ENSMUST00000222944.2
|
Hpcal1
|
hippocalcin-like 1 |
| chr9_+_106247943 | 2.34 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr19_-_21418215 | 2.33 |
ENSMUST00000237823.2
|
Gda
|
guanine deaminase |
| chr7_+_127845984 | 2.32 |
ENSMUST00000164710.8
ENSMUST00000070656.12 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr14_-_54647647 | 2.30 |
ENSMUST00000228488.2
ENSMUST00000195970.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr19_+_11382092 | 2.30 |
ENSMUST00000153546.8
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C |
| chr5_-_110987441 | 2.26 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr2_-_59778560 | 2.18 |
ENSMUST00000153136.2
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr2_-_91762119 | 2.14 |
ENSMUST00000069423.13
|
Mdk
|
midkine |
| chr17_+_34406762 | 2.13 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_+_3239868 | 2.11 |
ENSMUST00000094471.10
ENSMUST00000110043.8 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr18_-_35795233 | 2.09 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr9_-_54569128 | 2.08 |
ENSMUST00000034822.12
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr17_+_34406523 | 2.08 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr2_+_84867554 | 2.06 |
ENSMUST00000077798.13
|
Ssrp1
|
structure specific recognition protein 1 |
| chr2_+_76200299 | 2.02 |
ENSMUST00000046389.5
|
Rbm45
|
RNA binding motif protein 45 |
| chr11_-_41891111 | 1.98 |
ENSMUST00000109290.2
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr2_+_152950388 | 1.95 |
ENSMUST00000189688.2
ENSMUST00000109799.8 ENSMUST00000003370.14 |
Hck
|
hemopoietic cell kinase |
| chr5_-_123859153 | 1.95 |
ENSMUST00000196282.5
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr13_+_54722823 | 1.93 |
ENSMUST00000026988.11
|
Arl10
|
ADP-ribosylation factor-like 10 |
| chr11_+_80191692 | 1.92 |
ENSMUST00000017836.8
|
Rhbdl3
|
rhomboid like 3 |
| chr7_+_127846121 | 1.89 |
ENSMUST00000167965.8
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr13_-_63579497 | 1.88 |
ENSMUST00000160931.2
ENSMUST00000099444.10 ENSMUST00000220684.2 ENSMUST00000161977.8 ENSMUST00000163091.8 |
Fancc
|
Fanconi anemia, complementation group C |
| chr17_+_34824827 | 1.86 |
ENSMUST00000037489.15
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr2_+_157401998 | 1.85 |
ENSMUST00000153739.9
ENSMUST00000173595.2 ENSMUST00000109526.2 ENSMUST00000173839.2 ENSMUST00000173041.8 ENSMUST00000173793.8 ENSMUST00000172487.2 ENSMUST00000088484.6 |
Nnat
|
neuronatin |
| chr13_+_54722833 | 1.83 |
ENSMUST00000156024.2
|
Arl10
|
ADP-ribosylation factor-like 10 |
| chr3_+_88049633 | 1.83 |
ENSMUST00000001455.13
ENSMUST00000119251.8 |
Mef2d
|
myocyte enhancer factor 2D |
| chr9_+_32607301 | 1.79 |
ENSMUST00000034534.13
ENSMUST00000050797.14 ENSMUST00000184887.2 |
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chrX_+_154045439 | 1.74 |
ENSMUST00000026324.10
|
Acot9
|
acyl-CoA thioesterase 9 |
| chr12_-_70158348 | 1.73 |
ENSMUST00000220689.2
|
Nin
|
ninein |
| chr12_+_77285770 | 1.73 |
ENSMUST00000062804.8
|
Fut8
|
fucosyltransferase 8 |
| chr5_-_123859070 | 1.64 |
ENSMUST00000031376.12
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr4_-_43454561 | 1.64 |
ENSMUST00000107926.8
ENSMUST00000107925.8 |
Cd72
|
CD72 antigen |
| chr16_-_90807983 | 1.61 |
ENSMUST00000170853.8
ENSMUST00000130813.3 ENSMUST00000118390.10 |
Synj1
|
synaptojanin 1 |
| chr11_-_45846291 | 1.60 |
ENSMUST00000011398.13
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr5_+_117457126 | 1.60 |
ENSMUST00000111967.8
|
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr3_+_88049875 | 1.59 |
ENSMUST00000107558.9
ENSMUST00000107559.3 |
Mef2d
|
myocyte enhancer factor 2D |
| chr16_-_4376471 | 1.58 |
ENSMUST00000230875.2
|
Tfap4
|
transcription factor AP4 |
| chr9_+_113570506 | 1.57 |
ENSMUST00000213663.2
|
Clasp2
|
CLIP associating protein 2 |
| chr7_+_101034604 | 1.55 |
ENSMUST00000130016.8
ENSMUST00000134143.2 |
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr19_-_6593049 | 1.55 |
ENSMUST00000113451.9
|
Slc22a12
|
solute carrier family 22 (organic anion/cation transporter), member 12 |
| chr10_-_43934774 | 1.55 |
ENSMUST00000239010.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr16_-_19703014 | 1.54 |
ENSMUST00000100083.5
ENSMUST00000231564.2 |
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr17_+_17669082 | 1.51 |
ENSMUST00000140134.2
|
Lix1
|
limb and CNS expressed 1 |
| chr6_+_48604157 | 1.48 |
ENSMUST00000203088.3
ENSMUST00000204958.2 |
AI854703
|
expressed sequence AI854703 |
| chr2_+_29779750 | 1.47 |
ENSMUST00000113763.8
ENSMUST00000113757.8 ENSMUST00000113756.8 ENSMUST00000133233.8 ENSMUST00000113759.9 ENSMUST00000113755.8 ENSMUST00000137558.8 ENSMUST00000046571.14 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr10_+_79613083 | 1.46 |
ENSMUST00000020575.5
|
Fstl3
|
follistatin-like 3 |
| chr12_+_44375747 | 1.42 |
ENSMUST00000020939.16
ENSMUST00000220126.2 |
Nrcam
|
neuronal cell adhesion molecule |
| chr9_+_117888124 | 1.41 |
ENSMUST00000123690.2
|
Azi2
|
5-azacytidine induced gene 2 |
| chr1_-_133728779 | 1.39 |
ENSMUST00000143567.8
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr2_+_84867783 | 1.38 |
ENSMUST00000168266.8
ENSMUST00000130729.3 |
Ssrp1
|
structure specific recognition protein 1 |
| chr10_+_69370038 | 1.37 |
ENSMUST00000182439.8
ENSMUST00000092434.12 ENSMUST00000047061.13 ENSMUST00000092432.12 ENSMUST00000092431.12 ENSMUST00000054167.15 |
Ank3
|
ankyrin 3, epithelial |
| chr7_+_99825886 | 1.32 |
ENSMUST00000178946.9
|
Kcne3
|
potassium voltage-gated channel, Isk-related subfamily, gene 3 |
| chrX_-_58613428 | 1.31 |
ENSMUST00000119833.8
ENSMUST00000131319.8 |
Fgf13
|
fibroblast growth factor 13 |
| chr5_-_134485081 | 1.31 |
ENSMUST00000111244.5
|
Gtf2ird1
|
general transcription factor II I repeat domain-containing 1 |
| chr2_+_70392351 | 1.26 |
ENSMUST00000094934.11
|
Gad1
|
glutamate decarboxylase 1 |
| chr11_-_78136767 | 1.25 |
ENSMUST00000002121.5
|
Supt6
|
SPT6, histone chaperone and transcription elongation factor |
| chr15_-_51855073 | 1.25 |
ENSMUST00000022927.11
|
Rad21
|
RAD21 cohesin complex component |
| chr7_-_79989900 | 1.22 |
ENSMUST00000133728.8
|
Unc45a
|
unc-45 myosin chaperone A |
| chr7_+_140796559 | 1.19 |
ENSMUST00000148975.3
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chrX_+_100342749 | 1.18 |
ENSMUST00000118111.8
ENSMUST00000130555.8 ENSMUST00000151528.8 |
Nlgn3
|
neuroligin 3 |
| chr4_-_45320579 | 1.16 |
ENSMUST00000030003.10
|
Exosc3
|
exosome component 3 |
| chr5_+_117457356 | 1.14 |
ENSMUST00000086464.8
|
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr2_+_32425327 | 1.14 |
ENSMUST00000133512.2
ENSMUST00000048375.6 |
Fam102a
|
family with sequence similarity 102, member A |
| chr6_+_28423539 | 1.13 |
ENSMUST00000020717.12
ENSMUST00000169841.2 |
Arf5
|
ADP-ribosylation factor 5 |
| chr10_+_22520910 | 1.13 |
ENSMUST00000042261.5
|
Slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr11_-_76289888 | 1.13 |
ENSMUST00000021204.4
|
Nxn
|
nucleoredoxin |
| chr10_+_69369632 | 1.11 |
ENSMUST00000182155.8
ENSMUST00000183169.8 ENSMUST00000183148.8 |
Ank3
|
ankyrin 3, epithelial |
| chr13_+_109822527 | 1.11 |
ENSMUST00000074103.12
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr11_-_45845992 | 1.10 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr4_+_12140263 | 1.10 |
ENSMUST00000050069.9
ENSMUST00000069128.8 |
Rbm12b1
|
RNA binding motif protein 12 B1 |
| chr6_-_122587005 | 1.09 |
ENSMUST00000032211.5
|
Gdf3
|
growth differentiation factor 3 |
| chr10_+_69369590 | 1.08 |
ENSMUST00000182884.8
|
Ank3
|
ankyrin 3, epithelial |
| chr7_+_127865472 | 1.07 |
ENSMUST00000033045.11
|
Slc5a2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr10_-_34003933 | 1.06 |
ENSMUST00000062784.8
|
Calhm6
|
calcium homeostasis modulator family member 6 |
| chr2_+_181162278 | 1.06 |
ENSMUST00000072334.12
|
Dnajc5
|
DnaJ heat shock protein family (Hsp40) member C5 |
| chr8_+_55407872 | 1.05 |
ENSMUST00000033915.9
|
Gpm6a
|
glycoprotein m6a |
| chr7_+_127084283 | 1.03 |
ENSMUST00000048896.8
|
Fbrs
|
fibrosin |
| chr4_-_43454600 | 1.03 |
ENSMUST00000098105.4
ENSMUST00000098104.10 ENSMUST00000030179.11 |
Cd72
|
CD72 antigen |
| chrX_-_165293419 | 1.01 |
ENSMUST00000112188.8
|
Tceanc
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr7_+_140796537 | 1.00 |
ENSMUST00000141804.2
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr4_-_58553311 | 1.00 |
ENSMUST00000107571.8
ENSMUST00000055018.11 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr19_-_46321218 | 0.99 |
ENSMUST00000238062.2
|
Cuedc2
|
CUE domain containing 2 |
| chr6_-_52222776 | 0.98 |
ENSMUST00000048026.10
|
Hoxa11
|
homeobox A11 |
| chr9_-_96774719 | 0.98 |
ENSMUST00000154146.8
|
Pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr1_+_152830720 | 0.97 |
ENSMUST00000043313.15
ENSMUST00000186621.2 |
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr7_-_6334239 | 0.97 |
ENSMUST00000127658.2
ENSMUST00000062765.14 |
Zfp583
|
zinc finger protein 583 |
| chr9_+_113570580 | 0.96 |
ENSMUST00000216817.2
|
Clasp2
|
CLIP associating protein 2 |
| chr12_+_44375665 | 0.96 |
ENSMUST00000110748.4
|
Nrcam
|
neuronal cell adhesion molecule |
| chr2_-_152672535 | 0.96 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr3_-_115800989 | 0.93 |
ENSMUST00000067485.4
|
Slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr6_+_107506678 | 0.92 |
ENSMUST00000049285.10
|
Lrrn1
|
leucine rich repeat protein 1, neuronal |
| chr9_+_118335294 | 0.91 |
ENSMUST00000084820.6
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4 |
| chr7_-_118042772 | 0.91 |
ENSMUST00000081574.8
|
Syt17
|
synaptotagmin XVII |
| chr5_-_110927803 | 0.90 |
ENSMUST00000112426.8
|
Pus1
|
pseudouridine synthase 1 |
| chr2_+_181162326 | 0.90 |
ENSMUST00000108797.8
|
Dnajc5
|
DnaJ heat shock protein family (Hsp40) member C5 |
| chr4_-_117539431 | 0.89 |
ENSMUST00000102687.4
|
Dmap1
|
DNA methyltransferase 1-associated protein 1 |
| chr9_+_100956145 | 0.88 |
ENSMUST00000189616.2
|
Msl2
|
MSL complex subunit 2 |
| chr18_-_43506277 | 0.87 |
ENSMUST00000118043.8
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr11_-_41891359 | 0.87 |
ENSMUST00000070735.10
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr6_-_67014348 | 0.86 |
ENSMUST00000204369.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr1_-_155108455 | 0.85 |
ENSMUST00000035914.5
|
BC034090
|
cDNA sequence BC034090 |
| chr6_-_52195663 | 0.85 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7 |
| chr11_-_58692770 | 0.83 |
ENSMUST00000094156.12
ENSMUST00000239007.2 ENSMUST00000238886.2 ENSMUST00000060581.5 |
Fam183b
|
family with sequence similarity 183, member B |
| chr2_+_20742115 | 0.83 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr9_-_107482494 | 0.82 |
ENSMUST00000102529.10
|
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr4_+_11156411 | 0.77 |
ENSMUST00000029865.4
|
Trp53inp1
|
transformation related protein 53 inducible nuclear protein 1 |
| chr4_-_82939330 | 0.77 |
ENSMUST00000071708.12
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr4_-_58553553 | 0.76 |
ENSMUST00000107575.9
ENSMUST00000107574.8 ENSMUST00000147354.8 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr11_+_4085164 | 0.75 |
ENSMUST00000003677.11
ENSMUST00000145705.8 |
Rnf215
|
ring finger protein 215 |
| chr11_-_100244866 | 0.74 |
ENSMUST00000173630.8
|
Hap1
|
huntingtin-associated protein 1 |
| chr18_-_35795175 | 0.73 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr16_+_92295009 | 0.73 |
ENSMUST00000023670.4
|
Clic6
|
chloride intracellular channel 6 |
| chr7_+_5023552 | 0.73 |
ENSMUST00000208728.2
ENSMUST00000085427.6 |
Ccdc106
Zfp865
|
coiled-coil domain containing 106 zinc finger protein 865 |
| chr14_+_105496008 | 0.73 |
ENSMUST00000181969.8
|
Ndfip2
|
Nedd4 family interacting protein 2 |
| chr10_+_74896383 | 0.71 |
ENSMUST00000164107.3
|
Bcr
|
BCR activator of RhoGEF and GTPase |
| chr11_+_101066867 | 0.70 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1 |
| chr6_+_108190050 | 0.70 |
ENSMUST00000032192.9
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr3_-_94566107 | 0.67 |
ENSMUST00000196655.5
ENSMUST00000200407.2 ENSMUST00000006123.11 ENSMUST00000196733.5 |
Tuft1
|
tuftelin 1 |
| chr11_+_78136569 | 0.67 |
ENSMUST00000002133.9
|
Sdf2
|
stromal cell derived factor 2 |
| chr13_+_6598185 | 0.67 |
ENSMUST00000021611.10
ENSMUST00000222485.2 |
Pitrm1
|
pitrilysin metallepetidase 1 |
| chr6_-_67014191 | 0.66 |
ENSMUST00000204282.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr11_+_120612278 | 0.65 |
ENSMUST00000018156.12
|
Rac3
|
Rac family small GTPase 3 |
| chr2_+_29780122 | 0.65 |
ENSMUST00000113762.8
ENSMUST00000113765.8 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr9_+_110134241 | 0.64 |
ENSMUST00000199592.5
ENSMUST00000068071.12 |
Elp6
|
elongator acetyltransferase complex subunit 6 |
| chr11_+_120612369 | 0.63 |
ENSMUST00000142229.2
|
Rac3
|
Rac family small GTPase 3 |
| chr2_+_26220403 | 0.62 |
ENSMUST00000114134.9
ENSMUST00000127453.2 |
Gpsm1
|
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
| chrX_-_139857424 | 0.61 |
ENSMUST00000033805.15
ENSMUST00000112978.2 |
Psmd10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr8_+_121854566 | 0.59 |
ENSMUST00000181609.2
|
Foxl1
|
forkhead box L1 |
| chr4_+_74170165 | 0.59 |
ENSMUST00000030102.12
|
Kdm4c
|
lysine (K)-specific demethylase 4C |
| chr17_-_90217868 | 0.59 |
ENSMUST00000086423.6
|
Gm10184
|
predicted pseudogene 10184 |
| chr7_+_27198740 | 0.58 |
ENSMUST00000098644.9
ENSMUST00000108355.2 ENSMUST00000238936.2 |
Prx
|
periaxin |
| chr10_-_98962187 | 0.58 |
ENSMUST00000060761.7
|
Phxr2
|
per-hexamer repeat gene 2 |
| chrX_-_146337046 | 0.57 |
ENSMUST00000112819.9
ENSMUST00000136789.8 |
Lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr13_-_63579551 | 0.57 |
ENSMUST00000073029.13
|
Fancc
|
Fanconi anemia, complementation group C |
| chr8_+_85763534 | 0.56 |
ENSMUST00000093360.12
|
Tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chrX_+_135950334 | 0.56 |
ENSMUST00000047852.8
|
Fam199x
|
family with sequence similarity 199, X-linked |
| chr5_-_32903687 | 0.55 |
ENSMUST00000135248.2
|
Pisd
|
phosphatidylserine decarboxylase |
| chr9_-_107482462 | 0.54 |
ENSMUST00000194433.6
|
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr13_+_81931642 | 0.53 |
ENSMUST00000224574.2
|
Cetn3
|
centrin 3 |
| chr7_-_102143980 | 0.52 |
ENSMUST00000058750.4
|
Olfr545
|
olfactory receptor 545 |
| chr7_-_66915756 | 0.51 |
ENSMUST00000207715.2
|
Mef2a
|
myocyte enhancer factor 2A |
| chr13_+_81931196 | 0.50 |
ENSMUST00000022009.10
ENSMUST00000223793.2 |
Cetn3
|
centrin 3 |
| chr7_+_25016492 | 0.50 |
ENSMUST00000128119.2
|
Megf8
|
multiple EGF-like-domains 8 |
| chr5_-_32903615 | 0.47 |
ENSMUST00000120591.8
|
Pisd
|
phosphatidylserine decarboxylase |
| chr3_-_92393193 | 0.46 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr6_+_108190163 | 0.42 |
ENSMUST00000203615.3
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chrX_+_100342813 | 0.41 |
ENSMUST00000065858.3
|
Nlgn3
|
neuroligin 3 |
| chr14_+_105496147 | 0.39 |
ENSMUST00000138283.2
|
Ndfip2
|
Nedd4 family interacting protein 2 |
| chr10_+_69369854 | 0.39 |
ENSMUST00000182557.8
|
Ank3
|
ankyrin 3, epithelial |
| chr10_-_63926044 | 0.39 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr15_-_85466009 | 0.39 |
ENSMUST00000023015.15
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr4_-_147932817 | 0.38 |
ENSMUST00000105718.8
ENSMUST00000135798.2 |
Zfp933
|
zinc finger protein 933 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.8 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 1.4 | 4.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 1.2 | 3.5 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 1.0 | 6.1 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 1.0 | 4.0 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 1.0 | 2.9 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) neutrophil mediated killing of fungus(GO:0070947) |
| 0.9 | 6.0 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.8 | 6.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.8 | 4.7 | GO:0030421 | defecation(GO:0030421) |
| 0.7 | 3.0 | GO:0021941 | positive regulation of serotonin secretion(GO:0014064) negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.6 | 5.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.6 | 2.3 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.5 | 1.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.5 | 5.9 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.5 | 1.6 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 0.5 | 2.6 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.5 | 2.0 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.5 | 4.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.4 | 4.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.4 | 2.6 | GO:0032847 | regulation of nitrogen utilization(GO:0006808) positive regulation of neuron maturation(GO:0014042) nitrogen utilization(GO:0019740) regulation of cellular pH reduction(GO:0032847) |
| 0.4 | 1.7 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.4 | 4.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.4 | 4.0 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.4 | 6.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.3 | 2.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 1.3 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 4.0 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 1.2 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.3 | 1.4 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 1.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.3 | 0.8 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.3 | 2.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 1.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 1.7 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.2 | 2.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 1.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 0.6 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 0.6 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.2 | 5.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 1.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 1.8 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.2 | 2.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 2.9 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.2 | 2.7 | GO:0035898 | parathyroid hormone secretion(GO:0035898) |
| 0.2 | 1.0 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 | 0.9 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.7 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.1 | 1.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.5 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 1.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 1.6 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 1.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 2.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 1.9 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 2.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.3 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.1 | 4.7 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 1.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 1.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 2.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.5 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 2.4 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.4 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.6 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.7 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 0.4 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.7 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.2 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.1 | 1.0 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.1 | 0.9 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.4 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 3.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 1.0 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 1.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 9.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.9 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.4 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.1 | 3.9 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 4.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 3.5 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.6 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 2.5 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 2.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 3.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 2.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 1.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 3.9 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 5.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 1.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.9 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 1.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.7 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 2.1 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 2.6 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.3 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 1.0 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 1.2 | 6.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.0 | 4.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.9 | 10.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.8 | 2.5 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.5 | 4.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.4 | 4.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 3.0 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.3 | 1.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.3 | 5.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 2.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 3.4 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 1.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.1 | 4.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 2.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 5.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 2.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 13.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 2.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 2.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 2.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.2 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.1 | 1.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 4.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 1.6 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 4.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 5.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 2.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 2.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.9 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 3.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.0 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 2.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 2.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 10.1 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 4.3 | GO:0005819 | spindle(GO:0005819) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 1.0 | 4.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 1.0 | 6.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.0 | 3.0 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 1.0 | 4.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.9 | 2.7 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.7 | 4.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.7 | 6.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.7 | 6.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.6 | 7.3 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.4 | 4.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.4 | 10.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.4 | 2.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 1.7 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.3 | 2.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 1.6 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.3 | 3.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.3 | 2.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 1.0 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.3 | 5.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 0.7 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.2 | 1.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 2.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 2.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 4.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 8.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 2.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.7 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 1.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 1.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 2.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 2.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 3.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 5.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 13.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.4 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 0.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 5.5 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 1.6 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 4.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.5 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.1 | 1.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 1.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 2.9 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 2.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.1 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 0.7 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 1.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 2.3 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 4.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 5.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 5.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 6.8 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.3 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 1.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.2 | GO:0004527 | exonuclease activity(GO:0004527) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 2.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 8.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 2.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 6.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 3.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 5.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 5.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 7.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 3.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 4.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 8.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 6.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 11.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 4.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 2.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 3.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 5.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 5.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 3.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 1.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 5.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 4.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 3.7 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 2.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.1 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 0.6 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |