Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
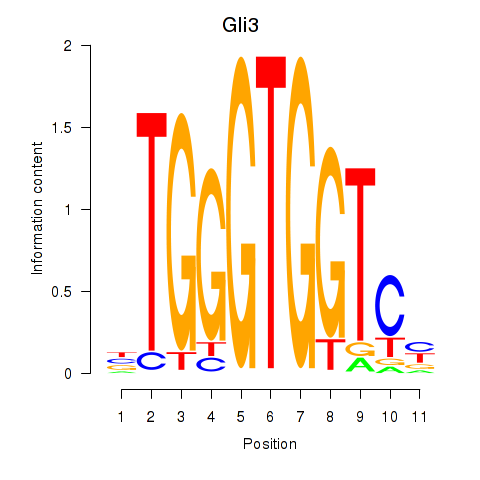
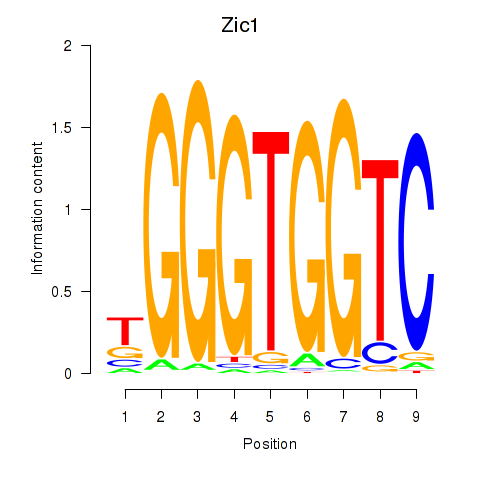
Results for Gli3_Zic1
Z-value: 1.98
Transcription factors associated with Gli3_Zic1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli3
|
ENSMUSG00000021318.16 | Gli3 |
|
Zic1
|
ENSMUSG00000032368.15 | Zic1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic1 | mm39_v1_chr9_-_91247809_91247827 | 0.71 | 4.0e-12 | Click! |
| Gli3 | mm39_v1_chr13_+_15638466_15638565 | 0.55 | 4.2e-07 | Click! |
Activity profile of Gli3_Zic1 motif
Sorted Z-values of Gli3_Zic1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gli3_Zic1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_157401998 | 27.76 |
ENSMUST00000153739.9
ENSMUST00000173595.2 ENSMUST00000109526.2 ENSMUST00000173839.2 ENSMUST00000173041.8 ENSMUST00000173793.8 ENSMUST00000172487.2 ENSMUST00000088484.6 |
Nnat
|
neuronatin |
| chr15_-_95426419 | 20.59 |
ENSMUST00000229933.2
ENSMUST00000166170.9 |
Nell2
|
NEL-like 2 |
| chr15_-_95426108 | 20.10 |
ENSMUST00000075275.3
|
Nell2
|
NEL-like 2 |
| chr7_-_100306160 | 19.98 |
ENSMUST00000107046.8
ENSMUST00000107045.9 ENSMUST00000139708.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chrX_+_100342749 | 18.64 |
ENSMUST00000118111.8
ENSMUST00000130555.8 ENSMUST00000151528.8 |
Nlgn3
|
neuroligin 3 |
| chr7_+_98917542 | 18.09 |
ENSMUST00000107100.3
ENSMUST00000208605.2 |
Map6
|
microtubule-associated protein 6 |
| chr11_+_101066867 | 16.80 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1 |
| chr4_-_68872585 | 16.19 |
ENSMUST00000030036.6
|
Brinp1
|
bone morphogenic protein/retinoic acid inducible neural specific 1 |
| chr11_-_41891359 | 15.16 |
ENSMUST00000070735.10
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr11_+_92989229 | 14.88 |
ENSMUST00000107859.8
ENSMUST00000107861.8 ENSMUST00000042943.13 ENSMUST00000107858.9 |
Car10
|
carbonic anhydrase 10 |
| chr7_-_74958121 | 14.86 |
ENSMUST00000085164.7
|
Sv2b
|
synaptic vesicle glycoprotein 2 b |
| chr8_+_120173458 | 13.66 |
ENSMUST00000098363.10
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr8_+_123844090 | 13.39 |
ENSMUST00000037900.9
|
Cpne7
|
copine VII |
| chrX_+_100342813 | 13.32 |
ENSMUST00000065858.3
|
Nlgn3
|
neuroligin 3 |
| chr3_+_68401536 | 13.19 |
ENSMUST00000182719.8
|
Schip1
|
schwannomin interacting protein 1 |
| chr9_+_50663171 | 13.19 |
ENSMUST00000214609.2
|
Cryab
|
crystallin, alpha B |
| chr7_+_98916635 | 12.72 |
ENSMUST00000122101.8
ENSMUST00000068973.11 ENSMUST00000207883.2 |
Map6
|
microtubule-associated protein 6 |
| chr19_-_38113249 | 12.57 |
ENSMUST00000112335.4
|
Rbp4
|
retinol binding protein 4, plasma |
| chr6_+_30738043 | 11.95 |
ENSMUST00000163949.9
ENSMUST00000124665.3 |
Mest
|
mesoderm specific transcript |
| chr11_-_41891111 | 11.43 |
ENSMUST00000109290.2
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr13_-_99653045 | 11.01 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr12_+_44375747 | 10.82 |
ENSMUST00000020939.16
ENSMUST00000220126.2 |
Nrcam
|
neuronal cell adhesion molecule |
| chr3_+_31204069 | 10.70 |
ENSMUST00000046174.8
|
Cldn11
|
claudin 11 |
| chr9_-_54569128 | 10.59 |
ENSMUST00000034822.12
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr8_-_65146079 | 10.56 |
ENSMUST00000048967.9
|
Cpe
|
carboxypeptidase E |
| chr7_-_118042772 | 10.07 |
ENSMUST00000081574.8
|
Syt17
|
synaptotagmin XVII |
| chr16_+_91066602 | 10.06 |
ENSMUST00000056882.7
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr12_+_44375665 | 9.90 |
ENSMUST00000110748.4
|
Nrcam
|
neuronal cell adhesion molecule |
| chr11_-_121279062 | 9.85 |
ENSMUST00000106107.3
|
Rab40b
|
Rab40B, member RAS oncogene family |
| chr10_-_63926044 | 9.72 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr6_-_124410452 | 9.56 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr19_-_38113696 | 9.38 |
ENSMUST00000025951.14
ENSMUST00000237287.2 |
Rbp4
|
retinol binding protein 4, plasma |
| chr7_+_98916677 | 9.29 |
ENSMUST00000127492.2
|
Map6
|
microtubule-associated protein 6 |
| chr11_-_42072990 | 9.26 |
ENSMUST00000205546.2
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr19_+_8641369 | 9.21 |
ENSMUST00000035444.10
ENSMUST00000163785.2 |
Chrm1
|
cholinergic receptor, muscarinic 1, CNS |
| chr3_+_45332831 | 9.02 |
ENSMUST00000193252.2
ENSMUST00000171554.8 ENSMUST00000166126.7 ENSMUST00000170695.4 |
Pcdh10
|
protocadherin 10 |
| chr19_+_4761181 | 8.91 |
ENSMUST00000008991.8
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr7_-_111379170 | 8.73 |
ENSMUST00000049430.15
ENSMUST00000106663.2 |
Galnt18
|
polypeptide N-acetylgalactosaminyltransferase 18 |
| chr15_-_85466009 | 8.72 |
ENSMUST00000023015.15
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr2_+_49509288 | 8.65 |
ENSMUST00000028102.14
|
Kif5c
|
kinesin family member 5C |
| chr8_+_55407872 | 8.59 |
ENSMUST00000033915.9
|
Gpm6a
|
glycoprotein m6a |
| chr1_+_50966670 | 8.46 |
ENSMUST00000081851.4
|
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr1_-_135095344 | 8.44 |
ENSMUST00000027682.9
|
Gpr37l1
|
G protein-coupled receptor 37-like 1 |
| chr7_-_142211203 | 8.29 |
ENSMUST00000097936.9
ENSMUST00000000033.12 |
Igf2
|
insulin-like growth factor 2 |
| chr11_+_98644340 | 8.20 |
ENSMUST00000124072.2
|
Thra
|
thyroid hormone receptor alpha |
| chr11_-_42072920 | 8.17 |
ENSMUST00000207274.2
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr11_+_103540391 | 8.13 |
ENSMUST00000057870.4
|
Rprml
|
reprimo-like |
| chr2_-_92264374 | 8.12 |
ENSMUST00000111278.2
ENSMUST00000090559.12 |
Cry2
|
cryptochrome 2 (photolyase-like) |
| chr3_-_95811993 | 8.04 |
ENSMUST00000147962.3
ENSMUST00000036181.15 |
Car14
|
carbonic anhydrase 14 |
| chrX_-_93585668 | 7.94 |
ENSMUST00000026142.8
|
Maged1
|
MAGE family member D1 |
| chr7_+_5059855 | 7.82 |
ENSMUST00000208161.2
ENSMUST00000207215.2 |
Ccdc106
|
coiled-coil domain containing 106 |
| chr5_-_118382926 | 7.78 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr7_+_5060159 | 7.72 |
ENSMUST00000045543.8
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr4_+_48585275 | 7.69 |
ENSMUST00000123476.8
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr11_-_96714813 | 7.57 |
ENSMUST00000142065.2
ENSMUST00000167110.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr7_+_5059703 | 7.54 |
ENSMUST00000208042.2
ENSMUST00000207974.2 |
Ccdc106
|
coiled-coil domain containing 106 |
| chr5_-_5315968 | 7.44 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr1_+_162466717 | 7.10 |
ENSMUST00000028020.11
|
Myoc
|
myocilin |
| chr6_-_28830344 | 7.09 |
ENSMUST00000171353.2
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr19_-_38113056 | 7.07 |
ENSMUST00000236283.2
|
Rbp4
|
retinol binding protein 4, plasma |
| chr6_-_115229128 | 7.04 |
ENSMUST00000032462.9
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr10_-_49664839 | 6.89 |
ENSMUST00000220263.2
ENSMUST00000218823.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr14_+_70768257 | 6.86 |
ENSMUST00000047331.8
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr2_+_70392351 | 6.78 |
ENSMUST00000094934.11
|
Gad1
|
glutamate decarboxylase 1 |
| chr1_-_134162231 | 6.65 |
ENSMUST00000169927.2
|
Adora1
|
adenosine A1 receptor |
| chr5_+_27022355 | 6.62 |
ENSMUST00000071500.13
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr6_-_119307685 | 6.61 |
ENSMUST00000112756.8
ENSMUST00000068351.14 |
Lrtm2
|
leucine-rich repeats and transmembrane domains 2 |
| chr5_+_30745447 | 6.57 |
ENSMUST00000066295.5
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr6_-_115228800 | 6.57 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr13_+_54519161 | 6.57 |
ENSMUST00000026985.9
|
Cplx2
|
complexin 2 |
| chrX_-_141173487 | 6.54 |
ENSMUST00000112904.8
ENSMUST00000112903.8 ENSMUST00000033634.5 |
Acsl4
|
acyl-CoA synthetase long-chain family member 4 |
| chr15_-_53765869 | 6.50 |
ENSMUST00000078673.14
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr16_+_20514925 | 6.48 |
ENSMUST00000128273.2
|
Fam131a
|
family with sequence similarity 131, member A |
| chrX_-_146337046 | 6.46 |
ENSMUST00000112819.9
ENSMUST00000136789.8 |
Lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr1_-_134163102 | 6.44 |
ENSMUST00000187631.2
ENSMUST00000038191.8 ENSMUST00000086465.6 |
Adora1
|
adenosine A1 receptor |
| chr5_+_93241385 | 6.42 |
ENSMUST00000201421.4
ENSMUST00000202415.4 ENSMUST00000202217.3 |
Septin11
|
septin 11 |
| chr9_+_106247943 | 6.41 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr16_-_20060053 | 6.40 |
ENSMUST00000040880.9
|
Map6d1
|
MAP6 domain containing 1 |
| chr11_-_70924288 | 6.37 |
ENSMUST00000238695.2
|
6330403K07Rik
|
RIKEN cDNA 6330403K07 gene |
| chr9_-_54568950 | 6.30 |
ENSMUST00000128624.2
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr14_+_70768289 | 6.27 |
ENSMUST00000226548.2
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr14_+_84680993 | 6.22 |
ENSMUST00000071370.7
|
Pcdh17
|
protocadherin 17 |
| chr19_+_4771089 | 6.16 |
ENSMUST00000238976.3
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr5_-_128897086 | 6.13 |
ENSMUST00000198941.5
ENSMUST00000199537.5 |
Rimbp2
|
RIMS binding protein 2 |
| chr12_+_95658987 | 6.12 |
ENSMUST00000057324.4
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr9_-_44646487 | 5.94 |
ENSMUST00000034611.15
|
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr5_+_14075281 | 5.80 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr19_-_6134703 | 5.78 |
ENSMUST00000161548.8
|
Zfpl1
|
zinc finger like protein 1 |
| chr9_+_27702243 | 5.76 |
ENSMUST00000115243.9
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr7_-_19504446 | 5.71 |
ENSMUST00000003061.14
|
Bcam
|
basal cell adhesion molecule |
| chr6_-_113508536 | 5.70 |
ENSMUST00000032425.7
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr17_-_13070780 | 5.68 |
ENSMUST00000162389.2
ENSMUST00000162119.8 ENSMUST00000159223.8 |
Mas1
|
MAS1 oncogene |
| chrX_+_7656225 | 5.67 |
ENSMUST00000136930.8
ENSMUST00000115675.9 ENSMUST00000101694.10 |
Gripap1
|
GRIP1 associated protein 1 |
| chr13_-_95170755 | 5.57 |
ENSMUST00000162670.8
|
Pde8b
|
phosphodiesterase 8B |
| chr10_+_106306122 | 5.56 |
ENSMUST00000029404.17
ENSMUST00000217854.2 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr1_-_125839897 | 5.54 |
ENSMUST00000159417.2
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr11_-_116545071 | 5.45 |
ENSMUST00000021166.6
|
Cygb
|
cytoglobin |
| chr6_+_110622533 | 5.41 |
ENSMUST00000071076.13
ENSMUST00000172951.2 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr7_+_4693759 | 5.36 |
ENSMUST00000048248.9
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr7_+_4693603 | 5.35 |
ENSMUST00000120836.8
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr11_+_87651359 | 5.33 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr10_+_40505985 | 5.30 |
ENSMUST00000019977.8
ENSMUST00000214102.2 ENSMUST00000213503.2 ENSMUST00000213442.2 ENSMUST00000216830.2 |
Ddo
|
D-aspartate oxidase |
| chr1_-_33946802 | 5.22 |
ENSMUST00000115161.8
ENSMUST00000129464.8 ENSMUST00000062289.11 |
Bend6
|
BEN domain containing 6 |
| chr12_+_109419371 | 5.21 |
ENSMUST00000109844.11
ENSMUST00000109842.9 ENSMUST00000109843.8 |
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr4_-_155103837 | 5.18 |
ENSMUST00000126098.2
ENSMUST00000176194.8 |
Plch2
|
phospholipase C, eta 2 |
| chr18_+_5593566 | 5.12 |
ENSMUST00000160910.2
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr16_-_30369378 | 5.12 |
ENSMUST00000140402.8
|
Tmem44
|
transmembrane protein 44 |
| chr2_+_106523532 | 5.10 |
ENSMUST00000111063.8
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr7_+_81824544 | 5.07 |
ENSMUST00000032874.14
|
Sh3gl3
|
SH3-domain GRB2-like 3 |
| chr4_+_42950367 | 5.06 |
ENSMUST00000084662.12
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr17_-_24428351 | 5.06 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr1_+_153776323 | 5.04 |
ENSMUST00000140685.4
ENSMUST00000139476.8 |
Glul
|
glutamate-ammonia ligase (glutamine synthetase) |
| chr6_+_86172196 | 4.99 |
ENSMUST00000032066.13
|
Tgfa
|
transforming growth factor alpha |
| chr9_+_50664288 | 4.95 |
ENSMUST00000214962.2
ENSMUST00000216755.2 |
Cryab
|
crystallin, alpha B |
| chr4_-_4138432 | 4.95 |
ENSMUST00000070375.8
|
Penk
|
preproenkephalin |
| chr9_+_50664207 | 4.89 |
ENSMUST00000034562.9
|
Cryab
|
crystallin, alpha B |
| chr4_+_127881786 | 4.85 |
ENSMUST00000184063.3
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chrX_+_7656251 | 4.85 |
ENSMUST00000140540.2
|
Gripap1
|
GRIP1 associated protein 1 |
| chr7_-_142453722 | 4.82 |
ENSMUST00000000219.10
|
Th
|
tyrosine hydroxylase |
| chr17_+_17669082 | 4.82 |
ENSMUST00000140134.2
|
Lix1
|
limb and CNS expressed 1 |
| chr3_+_94386385 | 4.77 |
ENSMUST00000199775.5
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr12_+_109419575 | 4.74 |
ENSMUST00000173539.8
ENSMUST00000109841.9 |
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr13_+_55097200 | 4.69 |
ENSMUST00000026994.14
ENSMUST00000109994.9 |
Unc5a
|
unc-5 netrin receptor A |
| chr14_+_47121487 | 4.65 |
ENSMUST00000137543.9
|
Samd4
|
sterile alpha motif domain containing 4 |
| chr15_-_91075933 | 4.64 |
ENSMUST00000069511.8
|
Abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr13_-_9815276 | 4.62 |
ENSMUST00000130151.8
|
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr3_-_26386609 | 4.59 |
ENSMUST00000193603.6
|
Nlgn1
|
neuroligin 1 |
| chr7_+_73040908 | 4.57 |
ENSMUST00000128471.2
ENSMUST00000139780.3 |
Rgma
|
repulsive guidance molecule family member A |
| chr17_+_37361523 | 4.56 |
ENSMUST00000172792.8
ENSMUST00000174347.2 |
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr13_-_36918424 | 4.55 |
ENSMUST00000037623.15
|
Nrn1
|
neuritin 1 |
| chr17_-_24424456 | 4.48 |
ENSMUST00000201583.2
ENSMUST00000202925.4 ENSMUST00000167791.9 ENSMUST00000201960.4 ENSMUST00000040474.11 ENSMUST00000201089.4 ENSMUST00000201301.4 ENSMUST00000201805.4 ENSMUST00000168410.9 ENSMUST00000097376.10 |
Tbc1d24
|
TBC1 domain family, member 24 |
| chr11_-_116249883 | 4.47 |
ENSMUST00000149147.8
|
Rnf157
|
ring finger protein 157 |
| chr4_+_123077286 | 4.46 |
ENSMUST00000126995.2
|
Hpcal4
|
hippocalcin-like 4 |
| chr6_-_35285045 | 4.45 |
ENSMUST00000031868.5
|
Slc13a4
|
solute carrier family 13 (sodium/sulfate symporters), member 4 |
| chr19_+_6434416 | 4.41 |
ENSMUST00000035269.15
ENSMUST00000113483.2 |
Pygm
|
muscle glycogen phosphorylase |
| chr2_-_77000936 | 4.40 |
ENSMUST00000164114.9
ENSMUST00000049544.14 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr5_+_35971767 | 4.39 |
ENSMUST00000114203.8
ENSMUST00000150146.2 |
Ablim2
|
actin-binding LIM protein 2 |
| chrX_+_72108393 | 4.39 |
ENSMUST00000060418.8
|
Pnma3
|
paraneoplastic antigen MA3 |
| chr14_+_55797934 | 4.37 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr3_+_145464413 | 4.37 |
ENSMUST00000029845.15
|
Ddah1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr7_-_62862261 | 4.36 |
ENSMUST00000032738.7
|
Chrna7
|
cholinergic receptor, nicotinic, alpha polypeptide 7 |
| chr11_+_72909811 | 4.35 |
ENSMUST00000092937.13
|
Camkk1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr7_-_45016138 | 4.34 |
ENSMUST00000211067.2
ENSMUST00000003961.16 |
Ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr5_+_71857261 | 4.31 |
ENSMUST00000031122.9
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 |
| chr10_-_5144699 | 4.29 |
ENSMUST00000215467.2
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr12_+_4284009 | 4.27 |
ENSMUST00000179139.3
|
Ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr2_+_119629995 | 4.22 |
ENSMUST00000028763.10
|
Tyro3
|
TYRO3 protein tyrosine kinase 3 |
| chr4_-_62126662 | 4.19 |
ENSMUST00000222050.2
ENSMUST00000068822.4 |
Zfp37
|
zinc finger protein 37 |
| chrX_-_141173330 | 4.19 |
ENSMUST00000112907.8
|
Acsl4
|
acyl-CoA synthetase long-chain family member 4 |
| chr1_+_72863641 | 4.17 |
ENSMUST00000047328.11
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr12_+_24548568 | 4.16 |
ENSMUST00000222710.2
|
Taf1b
|
TATA-box binding protein associated factor, RNA polymerase I, B |
| chr12_+_74044435 | 4.10 |
ENSMUST00000221220.2
|
Syt16
|
synaptotagmin XVI |
| chr6_-_101354858 | 4.02 |
ENSMUST00000075994.11
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr12_+_76451177 | 3.99 |
ENSMUST00000219555.2
|
Hspa2
|
heat shock protein 2 |
| chr9_+_40180726 | 3.99 |
ENSMUST00000171835.9
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr15_-_78602313 | 3.99 |
ENSMUST00000229441.2
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2 |
| chr16_-_85599994 | 3.96 |
ENSMUST00000023610.15
|
Adamts1
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 |
| chr5_-_125135434 | 3.95 |
ENSMUST00000134404.6
ENSMUST00000199561.2 |
Ncor2
|
nuclear receptor co-repressor 2 |
| chr11_-_35871300 | 3.94 |
ENSMUST00000018993.7
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr11_+_49792627 | 3.94 |
ENSMUST00000093141.12
ENSMUST00000093142.12 |
Rasgef1c
|
RasGEF domain family, member 1C |
| chr3_+_115801106 | 3.92 |
ENSMUST00000029575.12
ENSMUST00000106501.8 |
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr18_-_39062201 | 3.90 |
ENSMUST00000134864.2
|
Fgf1
|
fibroblast growth factor 1 |
| chr11_-_99313078 | 3.83 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr5_+_35971697 | 3.82 |
ENSMUST00000130233.8
|
Ablim2
|
actin-binding LIM protein 2 |
| chr9_+_120132962 | 3.81 |
ENSMUST00000048121.13
|
Myrip
|
myosin VIIA and Rab interacting protein |
| chr1_-_69147185 | 3.78 |
ENSMUST00000121473.8
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chr7_+_29483383 | 3.77 |
ENSMUST00000032803.12
ENSMUST00000122387.2 |
Zfp30
|
zinc finger protein 30 |
| chr13_-_9815350 | 3.75 |
ENSMUST00000110636.9
ENSMUST00000152725.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr2_-_29142965 | 3.74 |
ENSMUST00000155949.2
ENSMUST00000154682.8 ENSMUST00000028141.6 ENSMUST00000071201.5 |
6530402F18Rik
Ntng2
|
RIKEN cDNA 6530402F18 gene netrin G2 |
| chr16_-_90807983 | 3.70 |
ENSMUST00000170853.8
ENSMUST00000130813.3 ENSMUST00000118390.10 |
Synj1
|
synaptojanin 1 |
| chr11_+_53410552 | 3.70 |
ENSMUST00000108987.8
ENSMUST00000121334.8 ENSMUST00000117061.8 |
Septin8
|
septin 8 |
| chr2_-_32271833 | 3.69 |
ENSMUST00000146423.2
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene |
| chr19_+_45003304 | 3.68 |
ENSMUST00000039016.14
|
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chr12_+_109419454 | 3.68 |
ENSMUST00000109846.11
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr14_+_55798517 | 3.67 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr16_+_20551853 | 3.65 |
ENSMUST00000115423.8
ENSMUST00000007171.13 ENSMUST00000232646.2 |
Chrd
|
chordin |
| chr16_+_18595589 | 3.64 |
ENSMUST00000043577.3
|
Cldn5
|
claudin 5 |
| chr7_+_57240894 | 3.63 |
ENSMUST00000039697.14
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr13_+_118851214 | 3.60 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr11_-_58692770 | 3.57 |
ENSMUST00000094156.12
ENSMUST00000239007.2 ENSMUST00000238886.2 ENSMUST00000060581.5 |
Fam183b
|
family with sequence similarity 183, member B |
| chr1_-_51955054 | 3.55 |
ENSMUST00000018561.14
ENSMUST00000114537.9 |
Myo1b
|
myosin IB |
| chr1_-_75482975 | 3.51 |
ENSMUST00000113567.10
ENSMUST00000113565.3 |
Obsl1
|
obscurin-like 1 |
| chr14_+_55798362 | 3.49 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr15_-_43733389 | 3.46 |
ENSMUST00000067469.6
|
Tmem74
|
transmembrane protein 74 |
| chr7_+_26958150 | 3.45 |
ENSMUST00000079258.7
|
Numbl
|
numb-like |
| chr2_+_20727274 | 3.34 |
ENSMUST00000114607.8
|
Etl4
|
enhancer trap locus 4 |
| chr8_+_127790772 | 3.33 |
ENSMUST00000079777.12
ENSMUST00000160272.8 ENSMUST00000162907.8 ENSMUST00000162536.8 ENSMUST00000026921.13 ENSMUST00000162665.8 ENSMUST00000162602.8 ENSMUST00000160581.8 ENSMUST00000161355.8 ENSMUST00000162531.8 ENSMUST00000160766.8 ENSMUST00000159537.8 |
Pard3
|
par-3 family cell polarity regulator |
| chr4_+_42949814 | 3.32 |
ENSMUST00000037872.10
ENSMUST00000098112.9 |
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr3_+_136376440 | 3.31 |
ENSMUST00000056758.9
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr3_+_94385602 | 3.31 |
ENSMUST00000199884.5
ENSMUST00000198316.5 ENSMUST00000197558.5 |
Celf3
|
CUGBP, Elav-like family member 3 |
| chr4_-_62126598 | 3.31 |
ENSMUST00000221329.2
ENSMUST00000129511.3 |
Zfp37
|
zinc finger protein 37 |
| chr17_+_37356854 | 3.30 |
ENSMUST00000025338.16
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr19_+_42034231 | 3.28 |
ENSMUST00000172244.8
ENSMUST00000081714.5 |
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr19_-_5135510 | 3.28 |
ENSMUST00000140389.8
ENSMUST00000151413.2 ENSMUST00000077066.8 |
Tmem151a
|
transmembrane protein 151A |
| chr18_+_34973605 | 3.28 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2 |
| chr5_+_115417725 | 3.17 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr11_+_68986043 | 3.16 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr17_-_23818458 | 3.12 |
ENSMUST00000057029.5
|
Zfp13
|
zinc finger protein 13 |
| chr6_+_149310471 | 3.12 |
ENSMUST00000086829.11
ENSMUST00000111513.9 |
Bicd1
|
BICD cargo adaptor 1 |
| chr2_+_170573727 | 3.11 |
ENSMUST00000029075.5
|
Dok5
|
docking protein 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 32.0 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 6.5 | 32.6 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 4.6 | 13.7 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 4.4 | 13.1 | GO:0070256 | regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 3.5 | 10.6 | GO:0030070 | insulin processing(GO:0030070) |
| 3.1 | 3.1 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 2.9 | 8.7 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 2.8 | 27.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 2.7 | 8.1 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 2.4 | 56.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 2.4 | 40.7 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 2.4 | 7.1 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 2.2 | 8.7 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 2.1 | 10.7 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 2.1 | 6.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 2.1 | 6.4 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 2.1 | 16.8 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 2.1 | 23.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 2.1 | 8.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 2.0 | 6.0 | GO:0009073 | tyrosine biosynthetic process(GO:0006571) aromatic amino acid family biosynthetic process(GO:0009073) aromatic amino acid family biosynthetic process, prephenate pathway(GO:0009095) |
| 1.8 | 9.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.8 | 5.3 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 1.7 | 5.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 1.5 | 4.6 | GO:0098942 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 1.5 | 4.4 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 1.4 | 10.0 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 1.4 | 4.3 | GO:0071874 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 1.4 | 4.2 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 1.2 | 14.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 1.2 | 8.5 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 1.2 | 9.6 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 1.2 | 10.5 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 1.1 | 6.8 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.1 | 3.3 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 1.0 | 3.9 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 1.0 | 4.8 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 1.0 | 5.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.9 | 3.8 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.9 | 5.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.9 | 4.5 | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) |
| 0.8 | 10.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.8 | 3.3 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.8 | 5.6 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.8 | 5.6 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.8 | 3.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.7 | 8.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.7 | 17.9 | GO:0007614 | short-term memory(GO:0007614) |
| 0.7 | 2.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.7 | 2.1 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.7 | 3.9 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.6 | 4.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.6 | 4.5 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.6 | 13.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.6 | 13.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.6 | 4.3 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.6 | 11.0 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.6 | 3.7 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.6 | 5.4 | GO:0015671 | gas transport(GO:0015669) oxygen transport(GO:0015671) |
| 0.6 | 15.1 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.6 | 2.3 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.6 | 4.5 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.6 | 7.3 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.6 | 3.9 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.5 | 9.3 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.5 | 3.3 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.5 | 9.7 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.5 | 3.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.5 | 6.2 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.5 | 6.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.5 | 5.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.5 | 2.0 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.5 | 49.3 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.5 | 7.1 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.5 | 5.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.5 | 4.6 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.5 | 6.9 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.4 | 1.3 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.4 | 4.7 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 2.9 | GO:0009750 | response to fructose(GO:0009750) |
| 0.4 | 17.9 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.4 | 5.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.4 | 18.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.4 | 1.6 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.4 | 3.6 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.4 | 4.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.4 | 3.9 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.4 | 2.7 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.4 | 4.2 | GO:0060068 | vagina development(GO:0060068) |
| 0.4 | 2.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.4 | 5.1 | GO:0048752 | negative regulation of endothelial cell differentiation(GO:0045602) semicircular canal morphogenesis(GO:0048752) |
| 0.4 | 2.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.3 | 10.8 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.3 | 4.0 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.3 | 4.9 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.3 | 7.9 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.3 | 3.4 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.3 | 4.0 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.3 | 4.6 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.3 | 5.8 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.3 | 6.6 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.3 | 28.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.3 | 2.1 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.3 | 2.1 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.3 | 4.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 5.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.3 | 3.8 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.2 | 10.6 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 1.6 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 4.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 12.8 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.2 | 4.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 5.0 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.2 | 3.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 1.6 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 1.8 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 5.5 | GO:1903831 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.2 | 3.1 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.2 | 1.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.9 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 0.9 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 4.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 7.3 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.2 | 0.8 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 13.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.2 | 7.4 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.1 | 5.3 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 1.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.7 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 2.2 | GO:0060482 | bronchus development(GO:0060433) lobar bronchus development(GO:0060482) |
| 0.1 | 0.6 | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation(GO:0003198) |
| 0.1 | 3.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 4.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 4.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 1.7 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 8.6 | GO:0003407 | neural retina development(GO:0003407) |
| 0.1 | 0.4 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.4 | GO:0045918 | epithelial fluid transport(GO:0042045) negative regulation of cytolysis(GO:0045918) |
| 0.1 | 1.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.8 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 3.4 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.1 | 0.9 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.4 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.5 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 6.6 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.1 | 0.4 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 1.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.1 | 7.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 6.0 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.5 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.1 | 2.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 5.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 3.7 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 3.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 3.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 3.7 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 2.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 5.0 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.1 | 9.2 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 6.2 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.1 | 0.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 4.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 4.0 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.1 | 13.2 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.7 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.6 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.1 | 5.6 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 0.1 | GO:0003221 | tricuspid valve formation(GO:0003195) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.3 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.1 | 0.4 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.1 | 7.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 3.5 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.2 | GO:1903027 | asymmetric Golgi ribbon formation(GO:0090164) regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.3 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.3 | GO:1904355 | establishment of protein localization to telomere(GO:0070200) positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 1.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.3 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 5.7 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 2.1 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 4.8 | GO:0008366 | ensheathment of neurons(GO:0007272) axon ensheathment(GO:0008366) |
| 0.0 | 0.7 | GO:0019430 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.0 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 1.5 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 4.6 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 3.1 | GO:2000278 | regulation of DNA biosynthetic process(GO:2000278) |
| 0.0 | 1.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 5.7 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 2.0 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 1.6 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.7 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 2.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 1.8 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.5 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.0 | 2.1 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.1 | GO:1905035 | regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.0 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.7 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 10.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.9 | 52.0 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 1.7 | 13.8 | GO:0008091 | spectrin(GO:0008091) |
| 1.5 | 7.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 1.5 | 37.0 | GO:0098984 | neuron to neuron synapse(GO:0098984) |
| 1.4 | 4.2 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 1.3 | 10.5 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 1.3 | 6.6 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 1.1 | 3.4 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 1.0 | 4.8 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.9 | 37.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.9 | 17.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.9 | 6.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.8 | 10.7 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.8 | 20.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.7 | 14.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.7 | 27.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.7 | 7.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.7 | 7.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.5 | 12.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.5 | 1.5 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.5 | 3.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.5 | 2.8 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.5 | 11.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 5.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.4 | 3.1 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.4 | 5.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.4 | 4.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.4 | 4.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.3 | 1.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 3.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.3 | 10.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 6.4 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.3 | 10.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.3 | 4.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 1.0 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.3 | 2.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.2 | 2.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 2.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 2.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 1.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 38.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 48.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 2.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 1.9 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 4.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 15.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 3.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 2.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 4.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 9.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 0.9 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 5.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 3.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 3.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 4.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 25.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 1.0 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 1.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 44.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 0.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 6.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 47.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 0.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 1.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 3.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 4.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 2.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 2.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 2.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 3.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 6.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 3.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 5.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 1.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 3.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 2.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 41.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.9 | GO:0030016 | myofibril(GO:0030016) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.7 | 29.0 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 3.4 | 13.7 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 2.5 | 25.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 2.2 | 8.7 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 2.1 | 47.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 2.1 | 8.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.8 | 27.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 1.8 | 9.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.8 | 20.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.7 | 11.8 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 1.7 | 10.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.5 | 13.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.4 | 8.7 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 1.4 | 5.4 | GO:0004096 | catalase activity(GO:0004096) |
| 1.4 | 5.4 | GO:0070905 | serine binding(GO:0070905) |
| 1.3 | 5.3 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 1.3 | 5.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 1.3 | 3.9 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 1.3 | 6.4 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 1.2 | 5.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 1.2 | 11.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.2 | 26.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 1.2 | 4.8 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 1.1 | 6.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.0 | 3.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 1.0 | 7.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.0 | 4.0 | GO:0051381 | histamine binding(GO:0051381) |
| 1.0 | 6.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 1.0 | 2.9 | GO:0010428 | methyl-CpNpG binding(GO:0010428) |
| 0.9 | 3.6 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.9 | 10.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.9 | 4.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.8 | 3.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.7 | 5.7 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.7 | 2.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.6 | 4.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.6 | 13.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 4.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.6 | 2.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.6 | 5.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.6 | 12.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.6 | 3.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 2.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 10.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.5 | 4.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.5 | 3.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.5 | 4.0 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.5 | 3.0 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.5 | 6.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.5 | 3.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.5 | 4.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.5 | 2.8 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.5 | 14.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.4 | 8.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 2.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 3.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 3.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 4.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 8.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 43.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.4 | 2.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 1.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.3 | 1.7 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.3 | 1.0 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.3 | 4.6 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.3 | 6.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 3.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 1.6 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 5.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 0.9 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.3 | 2.9 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.3 | 5.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 7.1 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.3 | 12.4 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.3 | 2.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 6.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 29.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.2 | 1.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 3.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 2.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 0.8 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 5.2 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.2 | 6.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 1.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 12.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 3.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 0.6 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 5.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 1.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.2 | 2.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 6.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 2.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.2 | 4.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 1.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 2.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 3.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 3.1 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 12.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 45.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 3.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 5.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 7.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.7 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.4 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 4.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 2.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 4.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.4 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 2.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.0 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 2.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 3.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 9.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.3 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.1 | 7.8 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 0.7 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 3.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.0 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 2.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 5.4 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 1.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 7.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 5.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 2.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 8.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 1.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 8.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.7 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 11.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 72.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 17.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 5.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 13.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 2.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 10.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 8.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 6.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 7.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 8.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 3.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 5.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 19.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.2 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 5.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 5.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 19.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 2.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 52.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.9 | 47.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 12.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.7 | 15.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.6 | 14.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 21.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 11.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.4 | 11.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.4 | 6.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.3 | 8.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 6.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 8.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 5.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 14.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 9.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.3 | 13.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 4.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 5.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 4.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 5.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 11.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 13.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 16.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.2 | 4.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 5.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 5.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 5.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 2.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 6.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 3.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 3.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 4.1 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.1 | 4.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.9 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 5.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 5.8 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 1.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 2.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.3 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.4 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.5 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |