Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Glis2
Z-value: 1.08
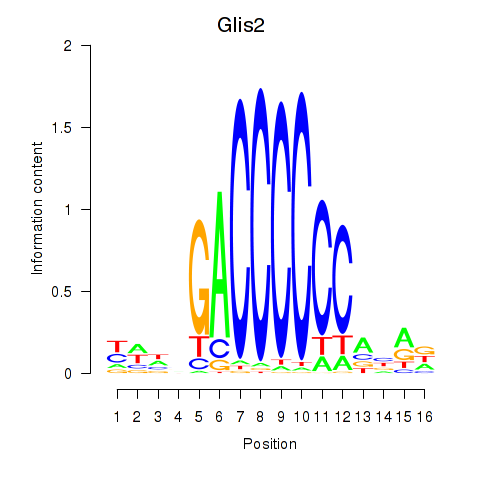
Transcription factors associated with Glis2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Glis2
|
ENSMUSG00000014303.14 | Glis2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Glis2 | mm39_v1_chr16_+_4412546_4412599 | 0.38 | 1.2e-03 | Click! |
Activity profile of Glis2 motif
Sorted Z-values of Glis2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Glis2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_75213570 | 11.19 |
ENSMUST00000213990.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr10_+_79650496 | 9.91 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr7_-_24705320 | 9.07 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr2_-_148574353 | 8.65 |
ENSMUST00000028926.13
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta |
| chr17_-_57394718 | 8.17 |
ENSMUST00000071135.6
|
Tubb4a
|
tubulin, beta 4A class IVA |
| chr6_-_115229128 | 7.52 |
ENSMUST00000032462.9
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr6_-_124410452 | 7.23 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr17_-_37334240 | 7.23 |
ENSMUST00000102665.11
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr16_-_45544960 | 7.00 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr5_-_38316296 | 6.97 |
ENSMUST00000201415.4
|
Nsg1
|
neuron specific gene family member 1 |
| chr4_-_149211145 | 6.76 |
ENSMUST00000030815.3
|
Cort
|
cortistatin |
| chr9_+_89791943 | 6.58 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr17_-_37334091 | 6.55 |
ENSMUST00000167275.3
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr13_+_58956077 | 6.33 |
ENSMUST00000109838.10
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr6_+_21215472 | 6.27 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chr11_+_103540391 | 6.22 |
ENSMUST00000057870.4
|
Rprml
|
reprimo-like |
| chr11_+_69909659 | 6.00 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr9_-_86762467 | 5.80 |
ENSMUST00000074501.12
ENSMUST00000239074.2 ENSMUST00000098495.10 ENSMUST00000036347.13 ENSMUST00000074468.13 |
Snap91
|
synaptosomal-associated protein 91 |
| chr10_+_126914755 | 5.30 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chrX_-_94240056 | 5.13 |
ENSMUST00000200628.2
ENSMUST00000197364.5 ENSMUST00000181987.8 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chr15_+_89384317 | 4.99 |
ENSMUST00000135214.2
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chrX_+_72546680 | 4.94 |
ENSMUST00000033744.12
ENSMUST00000088429.8 ENSMUST00000114479.2 |
Atp2b3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr16_-_44153498 | 4.89 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr16_+_29028447 | 4.81 |
ENSMUST00000089824.11
|
Plaat1
|
phospholipase A and acyltransferase 1 |
| chr7_+_28151370 | 4.45 |
ENSMUST00000190954.7
|
Lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr7_-_126303887 | 4.43 |
ENSMUST00000131415.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr11_+_97340962 | 4.36 |
ENSMUST00000107601.8
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr5_-_109704339 | 4.34 |
ENSMUST00000198960.2
|
Crlf2
|
cytokine receptor-like factor 2 |
| chr8_-_72178340 | 4.33 |
ENSMUST00000153800.8
ENSMUST00000146100.8 |
Fcho1
|
FCH domain only 1 |
| chrX_-_135769285 | 4.30 |
ENSMUST00000058814.7
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr17_-_52140305 | 4.22 |
ENSMUST00000133574.8
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr15_+_80171435 | 4.17 |
ENSMUST00000160424.8
|
Cacna1i
|
calcium channel, voltage-dependent, alpha 1I subunit |
| chr1_-_173195236 | 4.03 |
ENSMUST00000005470.5
ENSMUST00000111220.8 |
Cadm3
|
cell adhesion molecule 3 |
| chr7_+_45434755 | 3.90 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chrX_-_106446928 | 3.83 |
ENSMUST00000033591.6
|
Itm2a
|
integral membrane protein 2A |
| chrX_-_8072714 | 3.82 |
ENSMUST00000089403.10
ENSMUST00000077595.12 ENSMUST00000089402.10 ENSMUST00000082320.12 |
Porcn
|
porcupine O-acyltransferase |
| chr7_-_126303947 | 3.80 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chr10_-_25076008 | 3.76 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr2_+_164810139 | 3.73 |
ENSMUST00000202479.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr18_-_35841435 | 3.70 |
ENSMUST00000236738.2
ENSMUST00000237995.2 |
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr2_-_29142965 | 3.56 |
ENSMUST00000155949.2
ENSMUST00000154682.8 ENSMUST00000028141.6 ENSMUST00000071201.5 |
6530402F18Rik
Ntng2
|
RIKEN cDNA 6530402F18 gene netrin G2 |
| chr1_-_172156884 | 3.49 |
ENSMUST00000062387.8
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr9_-_97915036 | 3.41 |
ENSMUST00000162295.2
|
Clstn2
|
calsyntenin 2 |
| chr11_+_53410697 | 3.39 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr9_-_86762450 | 3.34 |
ENSMUST00000191290.3
|
Snap91
|
synaptosomal-associated protein 91 |
| chr5_-_49682106 | 3.34 |
ENSMUST00000176191.8
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr4_+_152160713 | 3.29 |
ENSMUST00000239025.2
|
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr13_+_43070127 | 3.28 |
ENSMUST00000239286.2
|
Phactr1
|
phosphatase and actin regulator 1 |
| chrX_-_151151680 | 3.22 |
ENSMUST00000070316.12
|
Gpr173
|
G-protein coupled receptor 173 |
| chr11_-_102837514 | 3.22 |
ENSMUST00000057849.6
|
C1ql1
|
complement component 1, q subcomponent-like 1 |
| chr4_-_41569500 | 3.22 |
ENSMUST00000108049.9
ENSMUST00000108052.10 ENSMUST00000108050.2 |
Fam219a
|
family with sequence similarity 219, member A |
| chr5_-_49682150 | 3.16 |
ENSMUST00000087395.11
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr17_-_23805187 | 3.14 |
ENSMUST00000227952.2
ENSMUST00000115516.11 |
Zfp13
|
zinc finger protein 13 |
| chr4_+_155819257 | 3.11 |
ENSMUST00000147721.8
ENSMUST00000127188.3 |
Tmem240
|
transmembrane protein 240 |
| chr6_-_90758954 | 3.06 |
ENSMUST00000238821.2
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr9_-_54554483 | 3.06 |
ENSMUST00000128163.8
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr3_+_129974531 | 3.00 |
ENSMUST00000080335.11
ENSMUST00000106353.2 |
Col25a1
|
collagen, type XXV, alpha 1 |
| chr8_+_105996419 | 3.00 |
ENSMUST00000036127.9
ENSMUST00000163734.9 |
Hsf4
|
heat shock transcription factor 4 |
| chr4_+_132948121 | 2.96 |
ENSMUST00000105910.2
|
Cd164l2
|
CD164 sialomucin-like 2 |
| chr1_-_172156828 | 2.93 |
ENSMUST00000194204.2
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr7_+_25016492 | 2.93 |
ENSMUST00000128119.2
|
Megf8
|
multiple EGF-like-domains 8 |
| chr2_-_57003064 | 2.91 |
ENSMUST00000112627.2
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr11_-_107805830 | 2.87 |
ENSMUST00000039071.3
|
Cacng5
|
calcium channel, voltage-dependent, gamma subunit 5 |
| chr5_+_30310138 | 2.87 |
ENSMUST00000058045.5
|
Garem2
|
GRB2 associated regulator of MAPK1 subtype 2 |
| chr2_-_80277969 | 2.83 |
ENSMUST00000028389.4
|
Frzb
|
frizzled-related protein |
| chr8_+_105996469 | 2.83 |
ENSMUST00000172525.8
ENSMUST00000174837.8 ENSMUST00000173859.2 |
Hsf4
|
heat shock transcription factor 4 |
| chr5_+_37495801 | 2.78 |
ENSMUST00000056365.9
|
Evc2
|
EvC ciliary complex subunit 2 |
| chr15_-_78428865 | 2.74 |
ENSMUST00000053239.4
|
Sstr3
|
somatostatin receptor 3 |
| chr10_+_84674008 | 2.64 |
ENSMUST00000095388.5
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr6_-_29212295 | 2.61 |
ENSMUST00000078155.12
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr5_-_30311073 | 2.54 |
ENSMUST00000199573.2
|
3110082J24Rik
|
RIKEN cDNA 3110082J24 gene |
| chr11_-_116226175 | 2.43 |
ENSMUST00000036215.8
|
Foxj1
|
forkhead box J1 |
| chrX_+_93278526 | 2.39 |
ENSMUST00000113908.8
ENSMUST00000113916.10 |
Klhl15
|
kelch-like 15 |
| chr19_+_45003304 | 2.37 |
ENSMUST00000039016.14
|
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chr7_+_5060159 | 2.36 |
ENSMUST00000045543.8
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr9_-_106343137 | 2.32 |
ENSMUST00000164834.3
|
Gpr62
|
G protein-coupled receptor 62 |
| chr4_-_128856213 | 2.28 |
ENSMUST00000119354.8
ENSMUST00000106068.8 ENSMUST00000030581.10 |
Azin2
|
antizyme inhibitor 2 |
| chr2_-_26184450 | 2.18 |
ENSMUST00000217256.2
ENSMUST00000227200.2 |
Ccdc187
|
coiled-coil domain containing 187 |
| chr12_+_72132621 | 2.11 |
ENSMUST00000057257.10
ENSMUST00000117449.8 |
Jkamp
|
JNK1/MAPK8-associated membrane protein |
| chr12_-_17061545 | 2.11 |
ENSMUST00000190691.2
|
2410004P03Rik
|
RIKEN cDNA 2410004P03 gene |
| chr7_-_114235506 | 2.08 |
ENSMUST00000205714.2
ENSMUST00000206853.2 ENSMUST00000205933.2 ENSMUST00000206156.2 ENSMUST00000032907.9 ENSMUST00000032906.11 |
Calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr11_+_4823951 | 2.08 |
ENSMUST00000038570.9
|
Nipsnap1
|
nipsnap homolog 1 |
| chr6_-_4747066 | 2.00 |
ENSMUST00000090686.11
ENSMUST00000133306.8 |
Sgce
|
sarcoglycan, epsilon |
| chr11_+_83599841 | 2.00 |
ENSMUST00000001009.14
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr7_+_5059855 | 1.97 |
ENSMUST00000208161.2
ENSMUST00000207215.2 |
Ccdc106
|
coiled-coil domain containing 106 |
| chr1_-_96799832 | 1.96 |
ENSMUST00000071985.6
|
Slco4c1
|
solute carrier organic anion transporter family, member 4C1 |
| chr6_-_116693849 | 1.96 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr6_-_29212239 | 1.95 |
ENSMUST00000160878.8
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr18_-_23174698 | 1.91 |
ENSMUST00000097651.10
|
Nol4
|
nucleolar protein 4 |
| chr16_+_17577493 | 1.88 |
ENSMUST00000165790.9
|
Klhl22
|
kelch-like 22 |
| chr11_+_69656797 | 1.86 |
ENSMUST00000108642.8
ENSMUST00000156932.8 |
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr16_+_29028860 | 1.80 |
ENSMUST00000162747.8
|
Plaat1
|
phospholipase A and acyltransferase 1 |
| chr7_-_126398343 | 1.79 |
ENSMUST00000032934.12
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr6_-_106777014 | 1.77 |
ENSMUST00000013882.10
ENSMUST00000113239.10 |
Crbn
|
cereblon |
| chr13_+_109769294 | 1.76 |
ENSMUST00000135275.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr16_+_17577464 | 1.75 |
ENSMUST00000129199.8
|
Klhl22
|
kelch-like 22 |
| chr9_-_121668527 | 1.73 |
ENSMUST00000135986.9
|
Ccdc13
|
coiled-coil domain containing 13 |
| chr5_-_137246852 | 1.69 |
ENSMUST00000179412.2
|
Muc3a
|
mucin 3A, cell surface associated |
| chr5_+_115417725 | 1.64 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr8_-_27903965 | 1.63 |
ENSMUST00000033882.10
|
Chrna6
|
cholinergic receptor, nicotinic, alpha polypeptide 6 |
| chr11_+_68989763 | 1.56 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1 |
| chr5_-_137246611 | 1.53 |
ENSMUST00000196391.5
|
Muc3a
|
mucin 3A, cell surface associated |
| chr17_+_34258411 | 1.52 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr2_+_105499233 | 1.52 |
ENSMUST00000111086.11
ENSMUST00000111087.10 |
Pax6
|
paired box 6 |
| chr7_-_44665639 | 1.46 |
ENSMUST00000085383.11
|
Scaf1
|
SR-related CTD-associated factor 1 |
| chr17_+_46961250 | 1.45 |
ENSMUST00000043464.14
|
Cul7
|
cullin 7 |
| chr8_+_73488496 | 1.45 |
ENSMUST00000058099.9
|
F2rl3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr16_+_72460029 | 1.43 |
ENSMUST00000023600.8
|
Robo1
|
roundabout guidance receptor 1 |
| chr4_-_49593875 | 1.42 |
ENSMUST00000151542.2
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chrX_+_149830166 | 1.40 |
ENSMUST00000026296.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr10_+_127102193 | 1.39 |
ENSMUST00000026479.11
|
Dctn2
|
dynactin 2 |
| chr14_+_55120875 | 1.38 |
ENSMUST00000134077.2
ENSMUST00000172844.8 ENSMUST00000133397.4 ENSMUST00000227108.2 |
Gm20521
Bcl2l2
|
predicted gene 20521 BCL2-like 2 |
| chr19_+_7245591 | 1.35 |
ENSMUST00000066646.12
|
Rcor2
|
REST corepressor 2 |
| chr2_+_119181703 | 1.34 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr16_+_17379749 | 1.30 |
ENSMUST00000171002.10
ENSMUST00000023441.11 |
P2rx6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr7_-_126398165 | 1.29 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr13_-_63712140 | 1.24 |
ENSMUST00000195756.6
|
Ptch1
|
patched 1 |
| chr11_+_59432388 | 1.23 |
ENSMUST00000079476.10
|
Nlrp3
|
NLR family, pyrin domain containing 3 |
| chr9_-_22218934 | 1.22 |
ENSMUST00000086278.7
ENSMUST00000215202.2 |
Zfp810
|
zinc finger protein 810 |
| chr15_+_41310815 | 1.21 |
ENSMUST00000090096.11
ENSMUST00000110297.10 |
Oxr1
|
oxidation resistance 1 |
| chr3_-_90416757 | 1.21 |
ENSMUST00000107343.8
ENSMUST00000001043.14 ENSMUST00000107344.8 ENSMUST00000076639.11 ENSMUST00000107346.8 ENSMUST00000146740.8 ENSMUST00000107342.2 ENSMUST00000049937.13 |
Chtop
|
chromatin target of PRMT1 |
| chr1_+_171954316 | 1.20 |
ENSMUST00000075895.9
ENSMUST00000111252.4 |
Pex19
|
peroxisomal biogenesis factor 19 |
| chr5_-_143255713 | 1.20 |
ENSMUST00000161448.8
|
Zfp316
|
zinc finger protein 316 |
| chr4_+_140737955 | 1.18 |
ENSMUST00000071977.9
|
Mfap2
|
microfibrillar-associated protein 2 |
| chr14_+_26836803 | 1.15 |
ENSMUST00000225949.2
|
Arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr4_-_117772163 | 1.09 |
ENSMUST00000036156.6
|
Ipo13
|
importin 13 |
| chr7_+_5023552 | 1.08 |
ENSMUST00000208728.2
ENSMUST00000085427.6 |
Ccdc106
Zfp865
|
coiled-coil domain containing 106 zinc finger protein 865 |
| chr4_-_62278761 | 1.07 |
ENSMUST00000107461.2
ENSMUST00000084528.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr17_-_65901946 | 1.06 |
ENSMUST00000232686.2
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr11_-_102120917 | 1.02 |
ENSMUST00000008999.12
|
Hdac5
|
histone deacetylase 5 |
| chr12_-_75224099 | 1.02 |
ENSMUST00000042299.4
|
Kcnh5
|
potassium voltage-gated channel, subfamily H (eag-related), member 5 |
| chr2_+_4980923 | 0.97 |
ENSMUST00000167607.8
ENSMUST00000115010.9 |
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr10_+_36850532 | 0.95 |
ENSMUST00000019911.14
ENSMUST00000105510.2 |
Hdac2
|
histone deacetylase 2 |
| chr2_+_153133326 | 0.94 |
ENSMUST00000028977.7
|
Kif3b
|
kinesin family member 3B |
| chr4_-_58499398 | 0.93 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr15_-_98465516 | 0.85 |
ENSMUST00000012104.7
|
Ccnt1
|
cyclin T1 |
| chr7_+_141056305 | 0.84 |
ENSMUST00000117634.2
|
Tspan4
|
tetraspanin 4 |
| chr12_-_31684588 | 0.83 |
ENSMUST00000020979.9
ENSMUST00000177962.9 |
Bcap29
|
B cell receptor associated protein 29 |
| chr13_-_76205115 | 0.83 |
ENSMUST00000056130.8
|
Gpr150
|
G protein-coupled receptor 150 |
| chr4_+_155875629 | 0.81 |
ENSMUST00000105593.2
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr11_+_101358990 | 0.80 |
ENSMUST00000001347.7
|
Rnd2
|
Rho family GTPase 2 |
| chr7_-_109322993 | 0.80 |
ENSMUST00000106735.9
ENSMUST00000033334.5 |
BC051019
|
cDNA sequence BC051019 |
| chr6_+_113581708 | 0.79 |
ENSMUST00000035725.7
|
Brk1
|
BRICK1, SCAR/WAVE actin-nucleating complex subunit |
| chr13_-_21726945 | 0.79 |
ENSMUST00000205976.3
ENSMUST00000175637.3 |
Olfr1366
|
olfactory receptor 1366 |
| chr7_+_141755098 | 0.78 |
ENSMUST00000187512.2
ENSMUST00000084414.6 |
Krtap5-3
|
keratin associated protein 5-3 |
| chr7_+_99030621 | 0.78 |
ENSMUST00000037528.10
|
Gdpd5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr8_-_25086976 | 0.77 |
ENSMUST00000033956.7
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr2_+_174171979 | 0.75 |
ENSMUST00000109083.2
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr12_-_11200306 | 0.75 |
ENSMUST00000055673.2
|
Kcns3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr10_-_81262948 | 0.75 |
ENSMUST00000078185.14
ENSMUST00000020461.15 ENSMUST00000105321.10 |
Nfic
|
nuclear factor I/C |
| chr11_+_74174562 | 0.75 |
ENSMUST00000214048.3
ENSMUST00000143976.4 ENSMUST00000205790.2 ENSMUST00000206659.2 |
Olfr59
|
olfactory receptor 59 |
| chr7_+_141765529 | 0.73 |
ENSMUST00000097943.2
|
Gm7579
|
predicted gene 7579 |
| chr2_-_152672535 | 0.72 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr2_+_4980986 | 0.70 |
ENSMUST00000027978.7
ENSMUST00000195688.2 |
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr6_+_48624295 | 0.69 |
ENSMUST00000078223.6
ENSMUST00000203509.2 |
Gimap8
|
GTPase, IMAP family member 8 |
| chr16_+_31918599 | 0.67 |
ENSMUST00000115168.9
|
Cep19
|
centrosomal protein 19 |
| chr7_+_127345909 | 0.66 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr8_-_23747057 | 0.65 |
ENSMUST00000051094.9
|
Golga7
|
golgi autoantigen, golgin subfamily a, 7 |
| chr19_+_6451667 | 0.65 |
ENSMUST00000113471.3
ENSMUST00000113469.3 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr2_+_112092271 | 0.65 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr17_-_31731222 | 0.64 |
ENSMUST00000236665.2
|
Wdr4
|
WD repeat domain 4 |
| chr16_+_16801246 | 0.64 |
ENSMUST00000232611.2
ENSMUST00000069107.14 |
Mapk1
|
mitogen-activated protein kinase 1 |
| chr7_+_44221791 | 0.64 |
ENSMUST00000002274.10
|
Napsa
|
napsin A aspartic peptidase |
| chr7_+_96981517 | 0.63 |
ENSMUST00000054107.6
|
Kctd21
|
potassium channel tetramerisation domain containing 21 |
| chr9_+_59587427 | 0.63 |
ENSMUST00000123914.8
|
Gramd2
|
GRAM domain containing 2 |
| chr19_-_58442866 | 0.62 |
ENSMUST00000169850.8
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr3_+_87855973 | 0.62 |
ENSMUST00000005019.6
|
Crabp2
|
cellular retinoic acid binding protein II |
| chr1_-_93729562 | 0.58 |
ENSMUST00000112890.3
|
Dtymk
|
deoxythymidylate kinase |
| chr2_-_26184563 | 0.56 |
ENSMUST00000057224.4
|
Ccdc187
|
coiled-coil domain containing 187 |
| chr15_-_25413784 | 0.56 |
ENSMUST00000228597.2
|
Basp1
|
brain abundant, membrane attached signal protein 1 |
| chr5_-_137531471 | 0.55 |
ENSMUST00000143495.8
ENSMUST00000111020.8 ENSMUST00000111023.8 ENSMUST00000111038.8 |
Gnb2
Epo
|
guanine nucleotide binding protein (G protein), beta 2 erythropoietin |
| chr1_-_134883577 | 0.54 |
ENSMUST00000168381.8
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr5_-_132570710 | 0.54 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr10_-_22607136 | 0.52 |
ENSMUST00000238910.2
ENSMUST00000127698.8 |
ENSMUSG00000118528.2
Tbpl1
|
novel protein TATA box binding protein-like 1 |
| chr5_-_137531413 | 0.51 |
ENSMUST00000168746.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr13_-_53083494 | 0.50 |
ENSMUST00000123599.8
|
Auh
|
AU RNA binding protein/enoyl-coenzyme A hydratase |
| chr19_-_40371242 | 0.49 |
ENSMUST00000224583.2
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr12_+_73170483 | 0.49 |
ENSMUST00000187549.7
ENSMUST00000021523.7 |
Mnat1
|
menage a trois 1 |
| chr19_+_45991907 | 0.48 |
ENSMUST00000099393.4
|
Hps6
|
HPS6, biogenesis of lysosomal organelles complex 2 subunit 3 |
| chr11_-_109613040 | 0.44 |
ENSMUST00000020938.8
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr12_-_17061504 | 0.44 |
ENSMUST00000189479.2
|
2410004P03Rik
|
RIKEN cDNA 2410004P03 gene |
| chr5_-_137531463 | 0.42 |
ENSMUST00000170293.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr7_-_4999042 | 0.41 |
ENSMUST00000162502.2
|
Zfp579
|
zinc finger protein 579 |
| chr19_+_53131187 | 0.40 |
ENSMUST00000050096.15
ENSMUST00000237832.2 |
Add3
|
adducin 3 (gamma) |
| chr4_-_62278673 | 0.40 |
ENSMUST00000084527.10
ENSMUST00000098033.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr7_+_101010447 | 0.39 |
ENSMUST00000137384.8
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr15_+_66763169 | 0.38 |
ENSMUST00000005255.9
|
Ccn4
|
cellular communication network factor 4 |
| chr1_-_118239146 | 0.35 |
ENSMUST00000027623.9
|
Tsn
|
translin |
| chr7_-_19398930 | 0.34 |
ENSMUST00000055242.11
|
Clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr6_-_148732893 | 0.33 |
ENSMUST00000145960.2
|
Ipo8
|
importin 8 |
| chr19_-_34618135 | 0.33 |
ENSMUST00000087357.5
|
Ifit1bl2
|
interferon induced protein with tetratricopeptide repeats 1B like 2 |
| chr7_-_23907518 | 0.33 |
ENSMUST00000086006.12
|
Zfp111
|
zinc finger protein 111 |
| chr8_+_3543131 | 0.31 |
ENSMUST00000061508.8
ENSMUST00000207318.2 |
Zfp358
|
zinc finger protein 358 |
| chr7_-_19595221 | 0.29 |
ENSMUST00000014830.8
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr4_+_155789545 | 0.29 |
ENSMUST00000105595.2
|
Ssu72
|
Ssu72 RNA polymerase II CTD phosphatase homolog (yeast) |
| chr13_-_13568106 | 0.28 |
ENSMUST00000021738.10
ENSMUST00000220628.2 |
Gpr137b
|
G protein-coupled receptor 137B |
| chr19_+_37538843 | 0.28 |
ENSMUST00000066439.8
ENSMUST00000238817.2 |
Exoc6
|
exocyst complex component 6 |
| chr14_-_8536869 | 0.27 |
ENSMUST00000238865.2
ENSMUST00000159275.4 |
Gm11100
|
predicted gene 11100 |
| chr6_-_54941673 | 0.26 |
ENSMUST00000203837.2
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr5_+_95488068 | 0.25 |
ENSMUST00000185432.2
|
Pramel47
|
PRAME like 47 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 9.9 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 1.5 | 9.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 1.4 | 4.3 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 1.2 | 3.7 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 1.1 | 6.3 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 1.0 | 8.2 | GO:0032796 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.8 | 2.4 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.8 | 3.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.7 | 2.9 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.7 | 2.9 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.7 | 5.0 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.7 | 4.9 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.7 | 7.0 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.7 | 2.1 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.7 | 4.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.6 | 3.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.6 | 3.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.6 | 7.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.6 | 8.7 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.6 | 9.1 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.6 | 2.3 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.6 | 2.8 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.6 | 11.2 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.6 | 7.2 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.5 | 3.8 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.5 | 6.0 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 0.5 | 1.4 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.4 | 4.9 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.4 | 2.6 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.4 | 3.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.4 | 8.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.3 | 2.4 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.3 | 1.2 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.3 | 5.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.3 | 0.8 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 0.8 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.3 | 1.1 | GO:1904937 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.3 | 1.6 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.3 | 0.5 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.3 | 0.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 0.9 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 | 1.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 4.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 4.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 3.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 10.9 | GO:0030431 | sleep(GO:0030431) |
| 0.2 | 6.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 1.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.6 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.2 | 1.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) fungiform papilla formation(GO:0061198) |
| 0.1 | 6.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 3.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.6 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 1.4 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 1.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.5 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 1.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.6 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 1.6 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.1 | 1.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 3.2 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 1.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.9 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 2.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 1.8 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 3.1 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.6 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 1.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 2.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 5.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 1.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.4 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.1 | 1.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 1.1 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 | 3.1 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 7.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 5.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:1903059 | N-terminal protein palmitoylation(GO:0006500) regulation of protein lipidation(GO:1903059) |
| 0.0 | 4.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 2.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.6 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 4.8 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 3.6 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 2.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.8 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 1.2 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 5.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.3 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 4.2 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 1.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.5 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 1.7 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.5 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 2.1 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 2.4 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.6 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.5 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 2.4 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 3.6 | GO:0060070 | canonical Wnt signaling pathway(GO:0060070) |
| 0.0 | 7.3 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.0 | 0.8 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.7 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.0 | 0.0 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.3 | GO:0033081 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 1.4 | 8.7 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.0 | 8.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.9 | 9.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.8 | 9.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.6 | 3.2 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.5 | 7.0 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.5 | 3.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.4 | 6.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 8.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 0.9 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.2 | 1.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 2.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 2.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 6.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 7.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 2.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 1.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.2 | 6.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 0.6 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 1.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 7.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 8.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.5 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.6 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 3.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 6.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 3.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.6 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 0.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 22.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.9 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 4.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 7.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 3.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 4.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 3.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 4.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 16.9 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 3.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.5 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 1.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 4.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 6.2 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 5.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 1.4 | 9.9 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 1.3 | 3.8 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 1.3 | 6.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.1 | 3.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 1.1 | 6.3 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 1.0 | 4.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.9 | 6.0 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.8 | 4.9 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.8 | 4.6 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.8 | 9.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.7 | 6.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.7 | 5.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.6 | 1.9 | GO:0010428 | methyl-CpNpG binding(GO:0010428) |
| 0.6 | 7.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.6 | 2.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.5 | 2.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.4 | 8.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 11.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.4 | 1.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 1.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.4 | 5.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 7.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 3.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 5.3 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.3 | 2.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.3 | 3.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 0.8 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.3 | 0.8 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.2 | 4.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 1.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.2 | 1.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 3.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 0.8 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 6.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 4.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 1.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 0.6 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 1.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.6 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 1.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 2.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 3.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 17.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 4.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 3.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 3.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 2.5 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 4.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 3.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 7.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 2.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 2.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 3.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 1.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 5.8 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 5.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.4 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 4.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 11.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 8.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 5.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 6.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 12.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 3.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 6.4 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 4.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 6.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 6.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 3.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 3.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 3.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 12.7 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.3 | 4.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 14.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 6.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 5.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 6.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 2.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 7.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.6 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 4.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 4.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 4.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 2.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.3 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 2.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.5 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |