Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Glis3
Z-value: 0.96
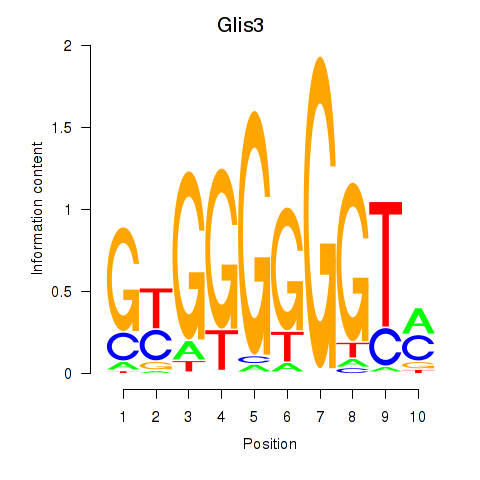
Transcription factors associated with Glis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Glis3
|
ENSMUSG00000052942.14 | Glis3 |
|
Glis3
|
ENSMUSG00000052942.14 | Glis3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Glis3 | mm39_v1_chr19_-_28657477_28657523 | 0.08 | 4.9e-01 | Click! |
Activity profile of Glis3 motif
Sorted Z-values of Glis3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Glis3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_113883285 | 7.56 |
ENSMUST00000090269.7
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr9_-_110571645 | 5.61 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr6_+_108637577 | 5.37 |
ENSMUST00000032194.11
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr9_-_50663648 | 5.20 |
ENSMUST00000217159.2
|
Hspb2
|
heat shock protein 2 |
| chr9_-_50663571 | 4.93 |
ENSMUST00000042790.5
|
Hspb2
|
heat shock protein 2 |
| chr11_-_106240215 | 4.87 |
ENSMUST00000021056.8
|
Scn4a
|
sodium channel, voltage-gated, type IV, alpha |
| chr7_+_44667377 | 4.66 |
ENSMUST00000044111.10
|
Rras
|
related RAS viral (r-ras) oncogene |
| chr7_-_19504446 | 4.52 |
ENSMUST00000003061.14
|
Bcam
|
basal cell adhesion molecule |
| chr2_+_74566740 | 4.32 |
ENSMUST00000111982.8
|
Hoxd3
|
homeobox D3 |
| chr17_-_24866749 | 4.20 |
ENSMUST00000234121.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr9_+_50663171 | 3.91 |
ENSMUST00000214609.2
|
Cryab
|
crystallin, alpha B |
| chr19_-_6964988 | 3.85 |
ENSMUST00000130048.8
ENSMUST00000025914.7 |
Vegfb
|
vascular endothelial growth factor B |
| chr2_+_32536594 | 3.79 |
ENSMUST00000113272.8
ENSMUST00000009705.14 ENSMUST00000167841.8 |
Eng
|
endoglin |
| chr1_-_168259264 | 3.37 |
ENSMUST00000176790.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr6_+_17463748 | 3.09 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr1_-_168259465 | 3.03 |
ENSMUST00000176540.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chrX_+_104807868 | 2.95 |
ENSMUST00000033581.4
|
Fgf16
|
fibroblast growth factor 16 |
| chr6_+_88175312 | 2.92 |
ENSMUST00000203480.2
ENSMUST00000015197.9 |
Gata2
|
GATA binding protein 2 |
| chr11_-_88609048 | 2.87 |
ENSMUST00000107909.8
|
Msi2
|
musashi RNA-binding protein 2 |
| chr1_-_168259710 | 2.79 |
ENSMUST00000072863.6
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chrX_-_23231245 | 2.75 |
ENSMUST00000115313.8
|
Klhl13
|
kelch-like 13 |
| chr14_+_55798517 | 2.73 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr4_+_144619397 | 2.71 |
ENSMUST00000105744.8
ENSMUST00000171001.8 |
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr11_+_96162283 | 2.70 |
ENSMUST00000000010.9
ENSMUST00000174042.3 |
Hoxb9
|
homeobox B9 |
| chr1_-_168259839 | 2.65 |
ENSMUST00000188912.7
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr9_-_48747232 | 2.58 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr14_+_55797934 | 2.54 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr6_-_52222776 | 2.49 |
ENSMUST00000048026.10
|
Hoxa11
|
homeobox A11 |
| chr6_+_17463819 | 2.45 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr4_-_120604445 | 2.42 |
ENSMUST00000030376.8
|
Kcnq4
|
potassium voltage-gated channel, subfamily Q, member 4 |
| chr11_-_88608958 | 2.40 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr1_-_168259070 | 2.37 |
ENSMUST00000064438.11
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr11_-_115310743 | 2.34 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr14_+_55798362 | 2.31 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr4_+_144619647 | 2.28 |
ENSMUST00000154208.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr6_+_17463925 | 2.27 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr4_+_144619696 | 2.24 |
ENSMUST00000142808.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr9_-_48747474 | 2.24 |
ENSMUST00000216150.2
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr2_-_75534985 | 2.23 |
ENSMUST00000102672.5
|
Nfe2l2
|
nuclear factor, erythroid derived 2, like 2 |
| chr4_+_119112590 | 2.14 |
ENSMUST00000084309.12
|
Cldn19
|
claudin 19 |
| chr9_-_108183356 | 2.09 |
ENSMUST00000192886.6
|
Tcta
|
T cell leukemia translocation altered gene |
| chr4_+_63133639 | 2.03 |
ENSMUST00000036300.13
|
Col27a1
|
collagen, type XXVII, alpha 1 |
| chr15_+_89383799 | 2.01 |
ENSMUST00000109309.9
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr6_+_108637816 | 1.99 |
ENSMUST00000163617.2
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr5_-_91550853 | 1.99 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr9_-_108183162 | 1.99 |
ENSMUST00000044725.9
|
Tcta
|
T cell leukemia translocation altered gene |
| chrM_+_8603 | 1.92 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr5_-_34345014 | 1.90 |
ENSMUST00000042701.13
ENSMUST00000119171.2 |
Mxd4
|
Max dimerization protein 4 |
| chr3_+_104696342 | 1.90 |
ENSMUST00000106787.8
ENSMUST00000176347.6 |
Rhoc
|
ras homolog family member C |
| chr4_+_119112692 | 1.85 |
ENSMUST00000094823.4
|
Cldn19
|
claudin 19 |
| chr7_+_27291126 | 1.83 |
ENSMUST00000167435.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr10_-_13350106 | 1.82 |
ENSMUST00000105545.12
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr9_-_108183140 | 1.81 |
ENSMUST00000195615.2
|
Tcta
|
T cell leukemia translocation altered gene |
| chr11_-_5848771 | 1.80 |
ENSMUST00000102921.4
|
Myl7
|
myosin, light polypeptide 7, regulatory |
| chr15_+_102314809 | 1.72 |
ENSMUST00000001326.7
|
Sp1
|
trans-acting transcription factor 1 |
| chr15_+_102314578 | 1.72 |
ENSMUST00000170884.8
ENSMUST00000163709.8 |
Sp1
|
trans-acting transcription factor 1 |
| chr3_+_104696108 | 1.71 |
ENSMUST00000002303.12
|
Rhoc
|
ras homolog family member C |
| chr6_-_72876686 | 1.70 |
ENSMUST00000206378.2
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr4_+_85972125 | 1.60 |
ENSMUST00000107178.9
ENSMUST00000048885.12 ENSMUST00000141889.8 ENSMUST00000120678.2 |
Adamtsl1
|
ADAMTS-like 1 |
| chr11_-_88608920 | 1.58 |
ENSMUST00000092794.12
|
Msi2
|
musashi RNA-binding protein 2 |
| chr7_+_27290969 | 1.57 |
ENSMUST00000108344.9
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr14_-_52151026 | 1.51 |
ENSMUST00000228164.2
|
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr2_-_160714473 | 1.50 |
ENSMUST00000103111.9
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr15_+_102829538 | 1.48 |
ENSMUST00000001700.7
|
Hoxc13
|
homeobox C13 |
| chr11_-_70128462 | 1.48 |
ENSMUST00000100950.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chrX_-_141749704 | 1.47 |
ENSMUST00000041317.3
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr7_-_49286594 | 1.46 |
ENSMUST00000032717.7
|
Dbx1
|
developing brain homeobox 1 |
| chr6_-_72876882 | 1.45 |
ENSMUST00000068697.11
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr3_+_68401536 | 1.44 |
ENSMUST00000182719.8
|
Schip1
|
schwannomin interacting protein 1 |
| chr16_+_22713593 | 1.42 |
ENSMUST00000232674.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr8_+_106002772 | 1.38 |
ENSMUST00000014920.8
|
Nol3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr13_+_119565118 | 1.34 |
ENSMUST00000109203.9
|
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr11_-_33097400 | 1.29 |
ENSMUST00000020507.8
|
Fgf18
|
fibroblast growth factor 18 |
| chr8_-_88686188 | 1.28 |
ENSMUST00000109655.9
|
Zfp423
|
zinc finger protein 423 |
| chr16_-_50252703 | 1.23 |
ENSMUST00000066037.13
ENSMUST00000089399.11 ENSMUST00000089404.10 ENSMUST00000138166.8 |
Bbx
|
bobby sox HMG box containing |
| chr4_+_118285275 | 1.23 |
ENSMUST00000006557.13
ENSMUST00000167636.8 ENSMUST00000102673.11 |
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr1_+_166829001 | 1.22 |
ENSMUST00000126198.3
|
Fam78b
|
family with sequence similarity 78, member B |
| chr8_+_23464860 | 1.21 |
ENSMUST00000110688.9
ENSMUST00000121802.9 |
Ank1
|
ankyrin 1, erythroid |
| chr3_+_121220146 | 1.13 |
ENSMUST00000029773.13
|
Cnn3
|
calponin 3, acidic |
| chr3_+_52175757 | 1.12 |
ENSMUST00000053764.7
|
Foxo1
|
forkhead box O1 |
| chr11_-_51891575 | 1.09 |
ENSMUST00000109086.8
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr10_-_120312374 | 1.08 |
ENSMUST00000072777.14
ENSMUST00000159699.2 |
Hmga2
|
high mobility group AT-hook 2 |
| chr11_-_51891259 | 1.07 |
ENSMUST00000020657.13
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr8_+_109441276 | 1.07 |
ENSMUST00000043896.10
|
Zfhx3
|
zinc finger homeobox 3 |
| chr2_+_4886298 | 1.05 |
ENSMUST00000027973.14
|
Sephs1
|
selenophosphate synthetase 1 |
| chr7_+_89779564 | 1.01 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr15_-_50753437 | 1.00 |
ENSMUST00000077935.6
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr19_-_47303184 | 0.98 |
ENSMUST00000237796.2
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chrX_+_100298134 | 0.96 |
ENSMUST00000062000.6
|
Foxo4
|
forkhead box O4 |
| chr2_-_160714904 | 0.95 |
ENSMUST00000109460.8
ENSMUST00000127201.2 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr3_-_142587419 | 0.94 |
ENSMUST00000174422.8
ENSMUST00000173830.8 |
Pkn2
|
protein kinase N2 |
| chr7_+_89779493 | 0.94 |
ENSMUST00000208730.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr16_+_20492014 | 0.94 |
ENSMUST00000154950.8
ENSMUST00000115461.8 ENSMUST00000136713.5 |
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr13_+_119565424 | 0.89 |
ENSMUST00000026520.14
|
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr7_-_46445305 | 0.89 |
ENSMUST00000107653.8
ENSMUST00000107654.8 ENSMUST00000014562.14 ENSMUST00000152759.8 |
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr6_+_85164420 | 0.88 |
ENSMUST00000045942.9
|
Emx1
|
empty spiracles homeobox 1 |
| chr9_+_118755521 | 0.88 |
ENSMUST00000073109.12
|
Ctdspl
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr7_+_79676095 | 0.87 |
ENSMUST00000206039.2
|
Zfp710
|
zinc finger protein 710 |
| chr11_+_100510043 | 0.86 |
ENSMUST00000107376.8
|
Nkiras2
|
NFKB inhibitor interacting Ras-like protein 2 |
| chr11_+_57692399 | 0.75 |
ENSMUST00000020826.6
|
Sap30l
|
SAP30-like |
| chr15_-_75760602 | 0.74 |
ENSMUST00000184858.2
|
Mroh6
|
maestro heat-like repeat family member 6 |
| chr17_-_85097945 | 0.72 |
ENSMUST00000112308.9
|
Lrpprc
|
leucine-rich PPR-motif containing |
| chr13_+_119565669 | 0.72 |
ENSMUST00000173627.8
ENSMUST00000126957.9 ENSMUST00000174691.8 |
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr11_-_72380730 | 0.71 |
ENSMUST00000045303.10
|
Spns2
|
spinster homolog 2 |
| chr9_+_95441652 | 0.71 |
ENSMUST00000079597.7
|
Paqr9
|
progestin and adipoQ receptor family member IX |
| chr10_+_63293284 | 0.69 |
ENSMUST00000105440.8
|
Ctnna3
|
catenin (cadherin associated protein), alpha 3 |
| chr7_-_46445085 | 0.68 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr1_-_84816379 | 0.64 |
ENSMUST00000187818.2
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr2_+_174171979 | 0.64 |
ENSMUST00000109083.2
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr2_-_160714749 | 0.62 |
ENSMUST00000176141.8
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr7_+_89779421 | 0.61 |
ENSMUST00000207225.2
ENSMUST00000207484.2 ENSMUST00000209068.2 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr13_-_103911092 | 0.61 |
ENSMUST00000074616.7
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr10_-_62723103 | 0.59 |
ENSMUST00000218438.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr13_-_40883893 | 0.59 |
ENSMUST00000021787.7
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr19_+_60878802 | 0.54 |
ENSMUST00000236876.2
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr17_-_34250616 | 0.54 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr13_+_38009981 | 0.54 |
ENSMUST00000110238.10
|
Rreb1
|
ras responsive element binding protein 1 |
| chr7_-_109271171 | 0.53 |
ENSMUST00000208734.2
|
Denn2b
|
DENN domain containing 2B |
| chr13_-_63721412 | 0.51 |
ENSMUST00000195106.2
|
Ptch1
|
patched 1 |
| chr11_-_115427007 | 0.50 |
ENSMUST00000118155.8
ENSMUST00000153892.2 |
Sumo2
|
small ubiquitin-like modifier 2 |
| chr14_+_53257873 | 0.47 |
ENSMUST00000196756.2
|
Trav7d-6
|
T cell receptor alpha variable 7D-6 |
| chr13_+_120151915 | 0.46 |
ENSMUST00000225543.2
|
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr9_+_21279802 | 0.45 |
ENSMUST00000214474.2
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr1_-_84817000 | 0.45 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr9_+_54672032 | 0.44 |
ENSMUST00000034830.9
|
Crabp1
|
cellular retinoic acid binding protein I |
| chr3_-_142587678 | 0.43 |
ENSMUST00000043812.15
|
Pkn2
|
protein kinase N2 |
| chr17_-_81035453 | 0.42 |
ENSMUST00000234133.2
ENSMUST00000112389.9 ENSMUST00000025089.9 |
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr5_-_21260878 | 0.42 |
ENSMUST00000030556.8
|
Ptpn12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr18_+_34910064 | 0.42 |
ENSMUST00000043775.9
ENSMUST00000224715.2 |
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chr15_+_27466732 | 0.42 |
ENSMUST00000022875.7
|
Ank
|
progressive ankylosis |
| chrX_-_103244728 | 0.40 |
ENSMUST00000056502.7
|
Nexmif
|
neurite extension and migration factor |
| chr10_-_68114543 | 0.39 |
ENSMUST00000219238.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr7_-_142211203 | 0.38 |
ENSMUST00000097936.9
ENSMUST00000000033.12 |
Igf2
|
insulin-like growth factor 2 |
| chrX_+_47430221 | 0.36 |
ENSMUST00000136348.8
|
Bcorl1
|
BCL6 co-repressor-like 1 |
| chr10_-_43934774 | 0.36 |
ENSMUST00000239010.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr19_+_8919228 | 0.35 |
ENSMUST00000096240.3
|
Mta2
|
metastasis-associated gene family, member 2 |
| chr16_+_20492267 | 0.33 |
ENSMUST00000115460.8
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr5_-_29683468 | 0.33 |
ENSMUST00000165512.4
ENSMUST00000001608.8 |
Mnx1
|
motor neuron and pancreas homeobox 1 |
| chr16_-_4376471 | 0.32 |
ENSMUST00000230875.2
|
Tfap4
|
transcription factor AP4 |
| chr1_+_166828982 | 0.31 |
ENSMUST00000165874.8
ENSMUST00000190081.7 |
Fam78b
|
family with sequence similarity 78, member B |
| chr7_+_18659787 | 0.31 |
ENSMUST00000032571.10
ENSMUST00000220302.2 |
Nova2
|
NOVA alternative splicing regulator 2 |
| chr17_-_34250474 | 0.30 |
ENSMUST00000171872.3
ENSMUST00000025186.16 |
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr15_+_89384317 | 0.30 |
ENSMUST00000135214.2
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chrX_-_93166992 | 0.29 |
ENSMUST00000088102.12
ENSMUST00000113927.8 |
Zfx
|
zinc finger protein X-linked |
| chrX_-_93166964 | 0.25 |
ENSMUST00000137853.8
|
Zfx
|
zinc finger protein X-linked |
| chr6_+_99669640 | 0.25 |
ENSMUST00000101122.3
|
Gpr27
|
G protein-coupled receptor 27 |
| chr11_+_115310963 | 0.21 |
ENSMUST00000106533.8
ENSMUST00000123345.2 |
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr9_+_106247943 | 0.21 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr7_+_46445512 | 0.21 |
ENSMUST00000006774.11
ENSMUST00000165031.8 |
Gtf2h1
|
general transcription factor II H, polypeptide 1 |
| chr7_+_126380655 | 0.20 |
ENSMUST00000172352.8
ENSMUST00000094037.5 |
Tbx6
|
T-box 6 |
| chrX_-_93585668 | 0.20 |
ENSMUST00000026142.8
|
Maged1
|
MAGE family member D1 |
| chr15_+_102315579 | 0.19 |
ENSMUST00000169619.2
|
Sp1
|
trans-acting transcription factor 1 |
| chr15_-_50753792 | 0.19 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr11_+_115310885 | 0.19 |
ENSMUST00000103035.10
|
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr9_+_106083988 | 0.18 |
ENSMUST00000188650.2
|
Twf2
|
twinfilin actin binding protein 2 |
| chr2_-_152673032 | 0.18 |
ENSMUST00000128172.3
|
Bcl2l1
|
BCL2-like 1 |
| chr13_+_38009951 | 0.17 |
ENSMUST00000138043.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr15_-_97902515 | 0.16 |
ENSMUST00000088355.12
|
Col2a1
|
collagen, type II, alpha 1 |
| chr17_+_35191661 | 0.15 |
ENSMUST00000007248.5
|
Hspa1l
|
heat shock protein 1-like |
| chr11_-_70128587 | 0.14 |
ENSMUST00000108576.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr4_-_133746138 | 0.14 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr3_+_89153258 | 0.13 |
ENSMUST00000040888.12
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr19_+_41471395 | 0.12 |
ENSMUST00000237208.2
ENSMUST00000238398.2 |
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr1_-_135615831 | 0.11 |
ENSMUST00000190298.8
|
Nav1
|
neuron navigator 1 |
| chr4_+_137004793 | 0.09 |
ENSMUST00000045747.5
|
Wnt4
|
wingless-type MMTV integration site family, member 4 |
| chr11_-_115426618 | 0.08 |
ENSMUST00000121185.8
ENSMUST00000117589.8 |
Sumo2
|
small ubiquitin-like modifier 2 |
| chr2_-_168584020 | 0.08 |
ENSMUST00000109177.8
|
Atp9a
|
ATPase, class II, type 9A |
| chr18_-_78249612 | 0.07 |
ENSMUST00000163367.3
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr9_+_107805647 | 0.07 |
ENSMUST00000085073.2
|
Actl11
|
actin-like 11 |
| chr15_-_97902576 | 0.06 |
ENSMUST00000023123.15
|
Col2a1
|
collagen, type II, alpha 1 |
| chr11_+_115921129 | 0.03 |
ENSMUST00000021116.12
ENSMUST00000106452.2 |
Unk
|
unkempt family zinc finger |
| chr7_+_5023375 | 0.03 |
ENSMUST00000076251.7
|
Zfp865
|
zinc finger protein 865 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0003032 | detection of oxygen(GO:0003032) cardiac jelly development(GO:1905072) |
| 1.2 | 3.6 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 1.1 | 7.8 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.9 | 10.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.8 | 3.8 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.7 | 2.2 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.7 | 2.9 | GO:0035854 | eosinophil fate commitment(GO:0035854) |
| 0.7 | 4.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.7 | 2.2 | GO:0033128 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.7 | 7.6 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.6 | 4.9 | GO:0015871 | choline transport(GO:0015871) |
| 0.6 | 4.2 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.6 | 3.4 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.5 | 4.8 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.5 | 6.9 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.5 | 2.4 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.5 | 1.4 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.4 | 2.6 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.4 | 14.2 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.4 | 3.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.3 | 7.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 | 1.5 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.3 | 5.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.3 | 2.0 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.3 | 4.0 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 2.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.2 | 1.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 6.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.2 | 1.5 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.2 | 1.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.2 | 0.7 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.2 | 2.7 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.2 | 3.6 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 0.4 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 3.4 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 1.0 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.7 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 0.6 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 1.1 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.1 | 1.5 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 2.0 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 1.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.5 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 6.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 1.5 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 2.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.6 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.7 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.8 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.4 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 1.4 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.0 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 1.2 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.2 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.4 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 1.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 1.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 1.9 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 1.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 4.8 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 0.6 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.5 | GO:0060746 | parental behavior(GO:0060746) |
| 0.0 | 4.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 2.7 | GO:0030879 | mammary gland development(GO:0030879) |
| 0.0 | 0.4 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.2 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.9 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.4 | 1.6 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.4 | 2.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 2.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 3.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 4.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 3.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 7.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 2.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 2.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 7.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 8.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 7.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 17.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 4.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 8.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 5.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 3.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 4.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.8 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 1.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 5.4 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 5.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 7.8 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.3 | 3.8 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.0 | 7.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.8 | 5.6 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.7 | 7.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.5 | 3.8 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 14.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 1.0 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.3 | 2.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 4.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.3 | 2.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 1.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 1.4 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.2 | 1.4 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.2 | 6.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 2.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 2.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 2.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 1.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.7 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 1.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 3.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.1 | 4.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 4.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 7.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 4.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.5 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.1 | 2.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 1.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.6 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 2.9 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 3.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 1.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 10.7 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 12.2 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 3.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 5.3 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 6.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 7.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 10.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 13.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 4.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 4.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 2.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 16.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.2 | 3.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 7.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 6.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 3.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 4.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 4.1 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.1 | 1.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 6.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 8.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |