Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Grhl1
Z-value: 3.34
Transcription factors associated with Grhl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Grhl1
|
ENSMUSG00000020656.17 | Grhl1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Grhl1 | mm39_v1_chr12_+_24622274_24622307 | 0.35 | 2.2e-03 | Click! |
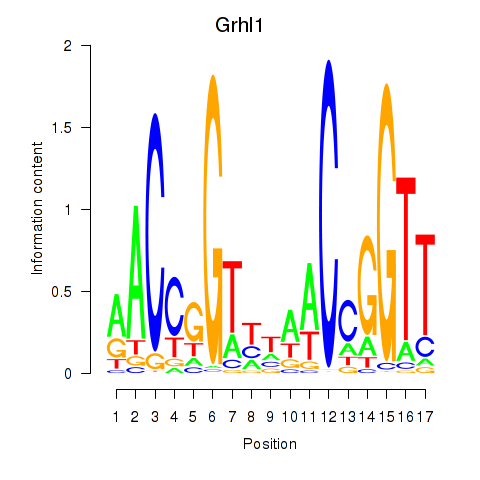
Activity profile of Grhl1 motif
Sorted Z-values of Grhl1 motif
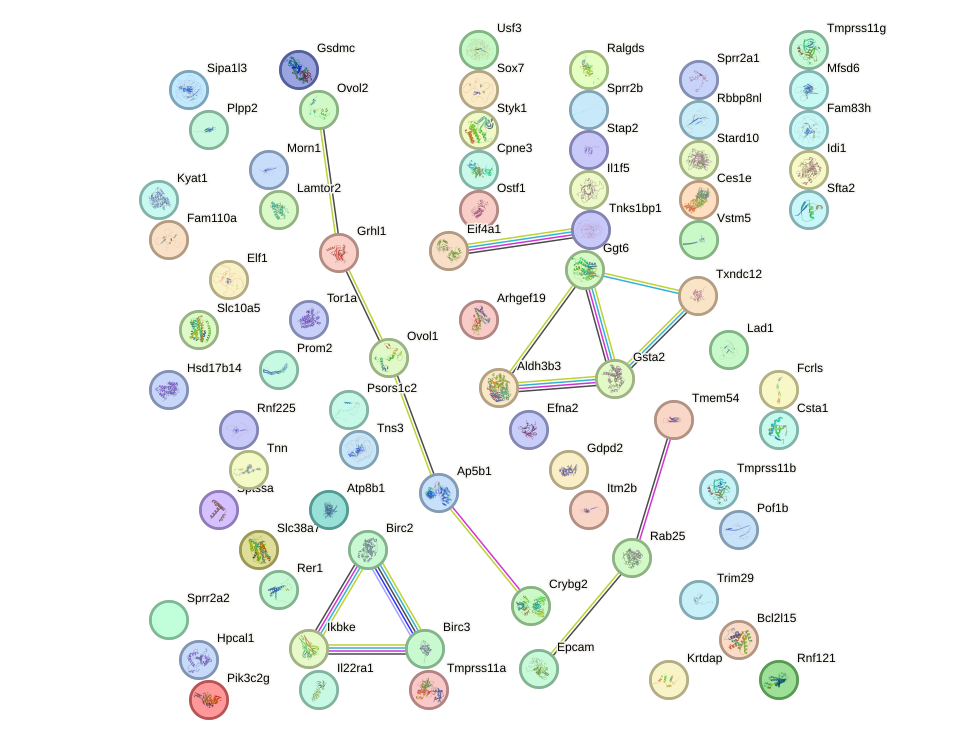
Network of associatons between targets according to the STRING database.
First level regulatory network of Grhl1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_88455556 | 62.06 |
ENSMUST00000131775.2
ENSMUST00000008745.13 |
Rab25
|
RAB25, member RAS oncogene family |
| chr3_+_92123106 | 44.91 |
ENSMUST00000074449.7
ENSMUST00000090871.3 |
Sprr2a1
Sprr2a2
|
small proline-rich protein 2A1 small proline-rich protein 2A2 |
| chrX_-_111608339 | 44.48 |
ENSMUST00000039887.4
|
Pof1b
|
premature ovarian failure 1B |
| chr7_+_30487322 | 42.11 |
ENSMUST00000189673.7
ENSMUST00000190990.7 ENSMUST00000189962.7 ENSMUST00000187493.7 ENSMUST00000098559.3 |
Krtdap
|
keratinocyte differentiation associated protein |
| chr3_+_92195873 | 41.74 |
ENSMUST00000090872.7
|
Sprr2a3
|
small proline-rich protein 2A3 |
| chr4_+_128999325 | 39.67 |
ENSMUST00000106064.10
ENSMUST00000030575.15 ENSMUST00000030577.11 |
Tmem54
|
transmembrane protein 54 |
| chr17_+_87943401 | 37.92 |
ENSMUST00000235125.2
ENSMUST00000053577.9 ENSMUST00000234009.2 |
Epcam
|
epithelial cell adhesion molecule |
| chr1_+_135746330 | 37.06 |
ENSMUST00000038760.10
|
Lad1
|
ladinin |
| chr9_+_78197205 | 35.54 |
ENSMUST00000119823.8
ENSMUST00000121273.2 |
Gsta5
|
glutathione S-transferase alpha 5 |
| chr18_-_64794338 | 34.68 |
ENSMUST00000025482.10
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr6_-_131293361 | 31.09 |
ENSMUST00000121078.2
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr15_-_63680596 | 31.07 |
ENSMUST00000110125.9
ENSMUST00000173503.3 |
Gsdmc
|
gasdermin C |
| chr9_+_43222104 | 28.48 |
ENSMUST00000034511.7
|
Trim29
|
tripartite motif-containing 29 |
| chr16_-_35951553 | 27.88 |
ENSMUST00000161638.2
ENSMUST00000096090.3 |
Csta1
|
cystatin A1 |
| chr2_-_127383305 | 25.68 |
ENSMUST00000103214.3
|
Prom2
|
prominin 2 |
| chr2_-_179931672 | 25.21 |
ENSMUST00000038529.2
|
Rbbp8nl
|
RBBP8 N-terminal like |
| chr17_-_56312555 | 24.15 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr2_+_24167275 | 24.11 |
ENSMUST00000114490.7
ENSMUST00000147885.4 |
Il1f5
|
interleukin 1 family, member 5 (delta) |
| chr2_-_127383332 | 23.85 |
ENSMUST00000028855.14
|
Prom2
|
prominin 2 |
| chr9_-_78254422 | 23.83 |
ENSMUST00000034902.12
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr2_+_24166920 | 23.52 |
ENSMUST00000168941.8
ENSMUST00000028360.8 ENSMUST00000123053.8 |
Il1f5
|
interleukin 1 family, member 5 (delta) |
| chr10_-_79369584 | 23.15 |
ENSMUST00000218241.2
ENSMUST00000166804.2 ENSMUST00000063879.13 |
Plpp2
|
phospholipid phosphatase 2 |
| chr8_-_93956143 | 22.46 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr19_-_5610628 | 22.04 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr9_-_78254443 | 21.74 |
ENSMUST00000129247.2
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr3_+_92223927 | 21.03 |
ENSMUST00000061038.4
|
Sprr2b
|
small proline-rich protein 2B |
| chr15_-_75886166 | 18.98 |
ENSMUST00000060807.12
|
Fam83h
|
family with sequence similarity 83, member H |
| chr17_+_35844091 | 18.66 |
ENSMUST00000025273.9
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 (human) |
| chr5_-_86616849 | 18.25 |
ENSMUST00000101073.3
|
Tmprss11a
|
transmembrane protease, serine 11a |
| chr11_+_72326391 | 17.39 |
ENSMUST00000100903.3
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr7_+_12661337 | 17.20 |
ENSMUST00000045870.5
|
Rnf225
|
ring finger protein 225 |
| chr3_-_10400710 | 16.85 |
ENSMUST00000078748.4
|
Slc10a5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5 |
| chr7_+_100970435 | 16.61 |
ENSMUST00000210192.2
ENSMUST00000172630.8 |
Stard10
|
START domain containing 10 |
| chr9_+_15150341 | 16.12 |
ENSMUST00000034413.8
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr7_+_100970910 | 15.44 |
ENSMUST00000174291.8
ENSMUST00000167888.9 ENSMUST00000172662.2 |
Stard10
|
START domain containing 10 |
| chr2_-_144174066 | 14.33 |
ENSMUST00000037423.4
|
Ovol2
|
ovo like zinc finger 2 |
| chr7_+_100971034 | 13.20 |
ENSMUST00000173270.8
|
Stard10
|
START domain containing 10 |
| chr4_+_135455427 | 13.00 |
ENSMUST00000102546.4
|
Il22ra1
|
interleukin 22 receptor, alpha 1 |
| chr4_+_140970161 | 12.88 |
ENSMUST00000138096.8
ENSMUST00000006618.9 ENSMUST00000125392.8 |
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr2_-_30095784 | 12.82 |
ENSMUST00000113662.8
|
Kyat1
|
kynurenine aminotransferase 1 |
| chr18_+_56565188 | 12.27 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr2_-_30095805 | 11.54 |
ENSMUST00000113663.9
ENSMUST00000044038.10 |
Kyat1
|
kynurenine aminotransferase 1 |
| chr5_-_86666408 | 11.18 |
ENSMUST00000140095.2
ENSMUST00000134179.8 |
Tmprss11g
|
transmembrane protease, serine 11g |
| chr4_+_112089442 | 10.70 |
ENSMUST00000038455.12
ENSMUST00000170945.2 |
Skint3
|
selection and upkeep of intraepithelial T cells 3 |
| chr4_+_133793208 | 10.16 |
ENSMUST00000137053.2
|
Crybg2
|
crystallin beta-gamma domain containing 2 |
| chr4_-_155170738 | 9.95 |
ENSMUST00000030914.4
|
Rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr7_+_45204317 | 9.60 |
ENSMUST00000107752.12
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr13_+_8935537 | 9.59 |
ENSMUST00000169314.9
|
Idi1
|
isopentenyl-diphosphate delta isomerase |
| chr1_-_159981132 | 9.34 |
ENSMUST00000039178.12
|
Tnn
|
tenascin N |
| chrX_-_37545311 | 9.20 |
ENSMUST00000074913.12
ENSMUST00000016678.14 ENSMUST00000061755.9 |
Lamp2
|
lysosomal-associated membrane protein 2 |
| chr2_-_151815307 | 7.87 |
ENSMUST00000109863.2
|
Fam110a
|
family with sequence similarity 110, member A |
| chr6_+_139576475 | 7.64 |
ENSMUST00000187618.7
ENSMUST00000186585.3 |
Pik3c2g
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr12_+_24622274 | 7.27 |
ENSMUST00000085553.13
|
Grhl1
|
grainyhead like transcription factor 1 |
| chr2_+_172392678 | 7.12 |
ENSMUST00000099058.10
|
Tfap2c
|
transcription factor AP-2, gamma |
| chr7_+_45204350 | 6.22 |
ENSMUST00000210300.2
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr7_-_29217967 | 5.94 |
ENSMUST00000181975.8
|
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr19_+_4008645 | 5.78 |
ENSMUST00000179433.8
|
Aldh3b3
|
aldehyde dehydrogenase 3 family, member B3 |
| chr1_-_131207279 | 5.65 |
ENSMUST00000062108.10
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr4_-_19570073 | 5.38 |
ENSMUST00000029885.5
|
Cpne3
|
copine III |
| chr2_+_84891281 | 5.33 |
ENSMUST00000238769.2
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr14_+_79718604 | 5.04 |
ENSMUST00000040131.13
|
Elf1
|
E74-like factor 1 |
| chr5_-_86824205 | 4.57 |
ENSMUST00000038448.7
|
Tmprss11b
|
transmembrane protease, serine 11B |
| chrX_+_99773784 | 4.34 |
ENSMUST00000113744.2
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr17_-_66191912 | 3.87 |
ENSMUST00000024905.11
|
Ralbp1
|
ralA binding protein 1 |
| chrX_+_99773523 | 3.73 |
ENSMUST00000019503.14
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr1_-_52766615 | 3.69 |
ENSMUST00000156876.8
ENSMUST00000087701.4 |
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr7_-_101714251 | 3.41 |
ENSMUST00000130074.2
ENSMUST00000131104.3 ENSMUST00000096639.12 |
Rnf121
|
ring finger protein 121 |
| chr18_-_46413886 | 3.35 |
ENSMUST00000236999.2
|
Pggt1b
|
protein geranylgeranyltransferase type I, beta subunit |
| chr11_-_69563133 | 3.19 |
ENSMUST00000163666.3
|
Eif4a1
|
eukaryotic translation initiation factor 4A1 |
| chr12_-_54703281 | 2.90 |
ENSMUST00000056228.8
|
Sptssa
|
serine palmitoyltransferase, small subunit A |
| chr17_+_35960600 | 2.81 |
ENSMUST00000171166.3
|
Sfta2
|
surfactant associated 2 |
| chr19_-_18609118 | 2.61 |
ENSMUST00000025631.7
ENSMUST00000236615.2 |
Ostf1
|
osteoclast stimulating factor 1 |
| chr5_+_124585244 | 2.48 |
ENSMUST00000198451.2
|
Kmt5a
|
lysine methyltransferase 5A |
| chr4_+_108691798 | 2.45 |
ENSMUST00000030296.9
|
Txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr12_+_17778198 | 2.36 |
ENSMUST00000222944.2
|
Hpcal1
|
hippocalcin-like 1 |
| chr11_-_8614667 | 2.01 |
ENSMUST00000239111.2
|
Tns3
|
tensin 3 |
| chr19_+_5618029 | 1.97 |
ENSMUST00000235575.2
ENSMUST00000235542.2 |
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr16_+_43993599 | 1.95 |
ENSMUST00000119746.8
ENSMUST00000088356.10 ENSMUST00000169582.3 |
Usf3
|
upstream transcription factor family member 3 |
| chr14_+_64181115 | 1.88 |
ENSMUST00000079652.7
|
Sox7
|
SRY (sex determining region Y)-box 7 |
| chr4_+_155171034 | 1.62 |
ENSMUST00000030915.11
ENSMUST00000155775.8 ENSMUST00000127457.2 |
Morn1
|
MORN repeat containing 1 |
| chr9_-_7873017 | 1.49 |
ENSMUST00000013949.15
|
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr10_-_128401773 | 1.46 |
ENSMUST00000026425.13
ENSMUST00000131728.4 |
Pa2g4
|
proliferation-associated 2G4 |
| chr2_-_30857858 | 1.42 |
ENSMUST00000028200.9
|
Tor1a
|
torsin family 1, member A (torsin A) |
| chr8_-_96580129 | 1.31 |
ENSMUST00000212628.2
ENSMUST00000040481.4 ENSMUST00000212270.2 |
Slc38a7
|
solute carrier family 38, member 7 |
| chr2_+_28403255 | 1.22 |
ENSMUST00000028170.15
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr3_+_103739877 | 0.86 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chr9_-_7873171 | 0.75 |
ENSMUST00000159323.2
ENSMUST00000115673.3 |
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr3_-_87171005 | 0.75 |
ENSMUST00000146512.2
|
Fcrls
|
Fc receptor-like S, scavenger receptor |
| chr14_-_73622638 | 0.71 |
ENSMUST00000228637.2
ENSMUST00000022704.9 |
Itm2b
|
integral membrane protein 2B |
| chr3_-_87170903 | 0.37 |
ENSMUST00000090986.11
|
Fcrls
|
Fc receptor-like S, scavenger receptor |
| chr19_+_18609291 | 0.18 |
ENSMUST00000042392.14
ENSMUST00000237347.2 |
Nmrk1
|
nicotinamide riboside kinase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.4 | 49.5 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 8.1 | 24.4 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 7.6 | 37.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 7.3 | 22.0 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 6.0 | 47.6 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 4.5 | 45.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 4.3 | 34.7 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 3.1 | 9.2 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 3.0 | 62.1 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 2.3 | 9.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 1.8 | 7.1 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 1.7 | 55.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 1.6 | 14.3 | GO:0060214 | endocardium formation(GO:0060214) |
| 1.5 | 107.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 1.2 | 17.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.9 | 5.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.8 | 3.9 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.7 | 9.9 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.6 | 44.5 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.6 | 7.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.6 | 31.1 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.5 | 15.8 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.5 | 8.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 23.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.4 | 3.4 | GO:0051771 | protein geranylgeranylation(GO:0018344) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.4 | 20.4 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.2 | 7.6 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.2 | 26.0 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.2 | 5.6 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.2 | 2.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 5.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.2 | 5.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 13.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 5.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.7 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 42.1 | GO:0008544 | epidermis development(GO:0008544) |
| 0.1 | 2.4 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 1.5 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.1 | 27.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 1.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 5.8 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.1 | 3.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 18.2 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.1 | 12.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.9 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.9 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 2.0 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 27.5 | GO:0006508 | proteolysis(GO:0006508) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.9 | 49.5 | GO:0044393 | microspike(GO:0044393) |
| 3.0 | 62.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 2.5 | 45.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 2.4 | 135.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 2.3 | 9.2 | GO:0097637 | platelet dense granule membrane(GO:0031088) intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 2.1 | 42.1 | GO:0042599 | lamellar body(GO:0042599) |
| 1.4 | 44.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.7 | 5.9 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.5 | 19.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 37.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.4 | 34.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.3 | 2.9 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 37.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 33.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 7.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 1.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 9.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 12.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 31.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 5.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 21.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 9.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 8.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 31.1 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 2.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 187.8 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 30.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 7.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.5 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 121.0 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 23.1 | GO:0005886 | plasma membrane(GO:0005886) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.9 | 47.6 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 8.1 | 24.4 | GO:0047945 | L-phenylalanine:pyruvate aminotransferase activity(GO:0047312) glutamine-phenylpyruvate transaminase activity(GO:0047316) L-glutamine:pyruvate aminotransferase activity(GO:0047945) |
| 4.3 | 13.0 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 3.5 | 34.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 2.7 | 19.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 2.6 | 66.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 2.3 | 15.8 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 2.2 | 62.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 2.1 | 16.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.9 | 9.6 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 1.6 | 45.6 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 1.5 | 23.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 1.4 | 17.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 1.1 | 22.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.9 | 5.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.8 | 2.4 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.8 | 8.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.8 | 5.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.6 | 5.8 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.6 | 49.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.6 | 2.9 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.6 | 3.4 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.4 | 7.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 31.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 1.3 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.3 | 9.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.3 | 44.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 29.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.9 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 34.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 99.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 36.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 3.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 29.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 12.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 5.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 9.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 7.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 41.4 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 3.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 19.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 7.2 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 2.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 5.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 7.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 9.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 81.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.6 | 34.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 7.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.3 | 9.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 5.6 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.3 | 8.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 9.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 7.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 3.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |