Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Gsc2_Dmbx1
Z-value: 0.49
Transcription factors associated with Gsc2_Dmbx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
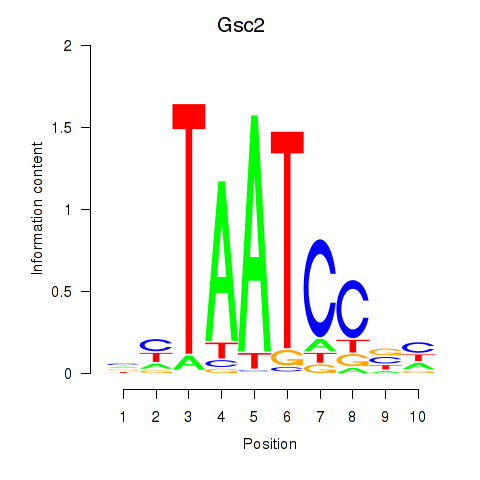
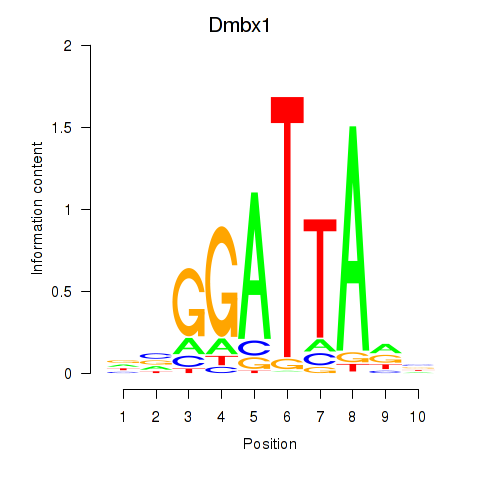
Gsc2
|
ENSMUSG00000022738.8 | Gsc2 |
|
Dmbx1
|
ENSMUSG00000028707.16 | Dmbx1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gsc2 | mm39_v1_chr16_-_17732923_17732923 | -0.43 | 2.0e-04 | Click! |
| Dmbx1 | mm39_v1_chr4_-_115797118_115797129 | -0.16 | 1.7e-01 | Click! |
Activity profile of Gsc2_Dmbx1 motif
Sorted Z-values of Gsc2_Dmbx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gsc2_Dmbx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_52173391 | 6.05 |
ENSMUST00000086844.10
|
Tcf7
|
transcription factor 7, T cell specific |
| chr6_+_119213468 | 4.74 |
ENSMUST00000238905.2
ENSMUST00000037434.13 |
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr6_+_119213541 | 2.96 |
ENSMUST00000186622.3
|
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr15_-_74606162 | 2.88 |
ENSMUST00000023260.5
|
Lypd2
|
Ly6/Plaur domain containing 2 |
| chr5_+_77413282 | 2.21 |
ENSMUST00000080359.12
|
Rest
|
RE1-silencing transcription factor |
| chr5_+_3393893 | 2.14 |
ENSMUST00000165117.8
ENSMUST00000197385.2 |
Cdk6
|
cyclin-dependent kinase 6 |
| chr2_+_101455079 | 2.11 |
ENSMUST00000111227.2
|
Rag2
|
recombination activating gene 2 |
| chr6_-_124817155 | 2.10 |
ENSMUST00000024206.6
|
Gnb3
|
guanine nucleotide binding protein (G protein), beta 3 |
| chr2_+_101455022 | 1.99 |
ENSMUST00000044031.4
|
Rag2
|
recombination activating gene 2 |
| chr5_+_3394187 | 1.94 |
ENSMUST00000042410.5
|
Cdk6
|
cyclin-dependent kinase 6 |
| chr14_+_28740162 | 1.73 |
ENSMUST00000055662.4
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr4_-_137493785 | 1.67 |
ENSMUST00000139951.8
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr11_+_90078485 | 1.61 |
ENSMUST00000124877.3
ENSMUST00000153734.2 ENSMUST00000134929.8 |
Gm45716
|
predicted gene 45716 |
| chr19_-_4189603 | 1.55 |
ENSMUST00000025761.8
|
Cabp4
|
calcium binding protein 4 |
| chr11_-_70405429 | 1.45 |
ENSMUST00000021179.4
|
Vmo1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr16_-_19703014 | 1.41 |
ENSMUST00000100083.5
ENSMUST00000231564.2 |
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr2_+_120307390 | 1.41 |
ENSMUST00000110716.9
ENSMUST00000028748.14 ENSMUST00000090028.13 ENSMUST00000110719.4 |
Capn3
|
calpain 3 |
| chr17_+_36172235 | 1.40 |
ENSMUST00000172931.2
|
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr14_+_26722319 | 1.32 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chrX_+_133486391 | 1.27 |
ENSMUST00000113211.8
|
Rpl36a
|
ribosomal protein L36A |
| chr11_-_70350783 | 1.25 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16 |
| chr7_-_4517367 | 1.18 |
ENSMUST00000166161.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr19_-_38032006 | 1.11 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr2_+_67276338 | 1.11 |
ENSMUST00000239060.2
ENSMUST00000028410.4 ENSMUST00000112347.8 ENSMUST00000238878.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr11_+_97917520 | 1.10 |
ENSMUST00000092425.11
|
Rpl19
|
ribosomal protein L19 |
| chr15_-_101801351 | 1.10 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr13_-_64645606 | 1.03 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr17_+_36172210 | 1.01 |
ENSMUST00000074259.15
ENSMUST00000174873.2 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr11_-_99176086 | 0.98 |
ENSMUST00000017255.4
|
Krt24
|
keratin 24 |
| chr19_+_4189786 | 0.96 |
ENSMUST00000096338.5
|
Gpr152
|
G protein-coupled receptor 152 |
| chr11_+_94901104 | 0.95 |
ENSMUST00000124735.2
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr8_+_13455080 | 0.95 |
ENSMUST00000033827.8
ENSMUST00000209909.2 |
Grk1
|
G protein-coupled receptor kinase 1 |
| chrX_+_159551171 | 0.95 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr9_-_50528530 | 0.93 |
ENSMUST00000147671.2
ENSMUST00000145139.8 ENSMUST00000155435.8 |
Nkapd1
|
NKAP domain containing 1 |
| chrX_+_73468140 | 0.93 |
ENSMUST00000135165.8
ENSMUST00000114128.8 ENSMUST00000004330.10 ENSMUST00000114133.9 ENSMUST00000114129.9 ENSMUST00000132749.2 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr5_-_144160397 | 0.93 |
ENSMUST00000085701.7
|
Tecpr1
|
tectonin beta-propeller repeat containing 1 |
| chrX_+_159551009 | 0.88 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr19_-_38031774 | 0.86 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr10_-_18890281 | 0.84 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr14_+_53100756 | 0.79 |
ENSMUST00000103616.5
ENSMUST00000186370.2 |
Trav15d-1-dv6d-1
|
T cell receptor alpha variable 15D-1-DV6D-1 |
| chr13_-_116446166 | 0.77 |
ENSMUST00000036060.13
|
Isl1
|
ISL1 transcription factor, LIM/homeodomain |
| chr9_+_30941924 | 0.76 |
ENSMUST00000216649.2
ENSMUST00000115222.10 |
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr4_+_80752535 | 0.72 |
ENSMUST00000102831.8
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr17_-_71309815 | 0.71 |
ENSMUST00000123686.8
|
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr8_+_79235946 | 0.69 |
ENSMUST00000209490.2
ENSMUST00000034111.10 |
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr2_+_104922363 | 0.68 |
ENSMUST00000111107.2
|
Gm11060
|
predicted gene 11060 |
| chr12_-_118265103 | 0.67 |
ENSMUST00000222314.2
ENSMUST00000026367.11 |
Sp4
|
trans-acting transcription factor 4 |
| chr4_+_80752360 | 0.66 |
ENSMUST00000133655.8
ENSMUST00000006151.13 |
Tyrp1
|
tyrosinase-related protein 1 |
| chr14_+_36776775 | 0.65 |
ENSMUST00000120052.2
|
Lrit1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr17_+_34092811 | 0.63 |
ENSMUST00000226885.2
|
Smim40-ps
|
small integral membrane protein 40, pseudogene |
| chr2_+_164404499 | 0.60 |
ENSMUST00000017867.10
ENSMUST00000109344.9 ENSMUST00000109345.9 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr3_-_103645311 | 0.56 |
ENSMUST00000029440.10
|
Olfml3
|
olfactomedin-like 3 |
| chr1_+_178014983 | 0.56 |
ENSMUST00000161075.8
ENSMUST00000027783.14 |
Desi2
|
desumoylating isopeptidase 2 |
| chr17_+_34124078 | 0.55 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr7_+_13132075 | 0.52 |
ENSMUST00000117400.2
|
Cabp5
|
calcium binding protein 5 |
| chr11_+_109540201 | 0.52 |
ENSMUST00000106677.8
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr17_-_28584117 | 0.51 |
ENSMUST00000233420.2
|
Tulp1
|
tubby like protein 1 |
| chr7_+_101619019 | 0.51 |
ENSMUST00000084852.13
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr5_-_137643954 | 0.51 |
ENSMUST00000057314.4
|
Irs3
|
insulin receptor substrate 3 |
| chr2_-_9888804 | 0.49 |
ENSMUST00000114915.3
|
9230102O04Rik
|
RIKEN cDNA 9230102O04 gene |
| chr6_+_3993774 | 0.48 |
ENSMUST00000031673.7
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr1_-_44258112 | 0.47 |
ENSMUST00000054801.4
|
Mettl21e
|
methyltransferase like 21E |
| chr4_+_136349924 | 0.46 |
ENSMUST00000178843.3
|
Lactbl1
|
lactamase, beta-like 1 |
| chr11_+_117090465 | 0.46 |
ENSMUST00000093907.11
|
Septin9
|
septin 9 |
| chr4_+_102446883 | 0.46 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr19_-_12742811 | 0.45 |
ENSMUST00000112933.2
|
Cntf
|
ciliary neurotrophic factor |
| chr17_+_28259749 | 0.45 |
ENSMUST00000233869.2
|
Anks1
|
ankyrin repeat and SAM domain containing 1 |
| chr14_+_54214610 | 0.45 |
ENSMUST00000199137.2
|
Trav23
|
T cell receptor alpha variable 23 |
| chr17_-_23892798 | 0.43 |
ENSMUST00000047436.11
ENSMUST00000115490.9 ENSMUST00000095579.11 |
Thoc6
|
THO complex 6 |
| chr1_+_12762501 | 0.43 |
ENSMUST00000177608.8
ENSMUST00000180062.8 |
Sulf1
|
sulfatase 1 |
| chr1_+_177272215 | 0.42 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr13_+_23936250 | 0.42 |
ENSMUST00000091703.3
|
H3c2
|
H3 clustered histone 2 |
| chr10_-_6930376 | 0.41 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr13_-_21967540 | 0.41 |
ENSMUST00000189457.2
|
H3c11
|
H3 clustered histone 11 |
| chr1_+_12762712 | 0.40 |
ENSMUST00000186051.7
|
Sulf1
|
sulfatase 1 |
| chr3_-_64197130 | 0.40 |
ENSMUST00000233531.2
ENSMUST00000176328.9 |
Vmn2r3
|
vomeronasal 2, receptor 3 |
| chrX_+_37689503 | 0.40 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr2_-_51862941 | 0.39 |
ENSMUST00000145481.8
ENSMUST00000112705.9 |
Nmi
|
N-myc (and STAT) interactor |
| chr11_-_107228382 | 0.39 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr8_-_106863521 | 0.38 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr9_+_35332837 | 0.38 |
ENSMUST00000119129.10
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr11_+_19874354 | 0.38 |
ENSMUST00000093299.13
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chrX_+_168662592 | 0.37 |
ENSMUST00000112105.8
ENSMUST00000078947.12 |
Mid1
|
midline 1 |
| chr9_+_108367801 | 0.36 |
ENSMUST00000006854.13
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr8_-_106863423 | 0.36 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr11_+_23206565 | 0.36 |
ENSMUST00000136235.2
|
Xpo1
|
exportin 1 |
| chr2_-_93292257 | 0.36 |
ENSMUST00000150508.8
|
Cd82
|
CD82 antigen |
| chr5_-_137643799 | 0.35 |
ENSMUST00000196511.2
|
Irs3
|
insulin receptor substrate 3 |
| chr14_-_55163452 | 0.35 |
ENSMUST00000227037.2
|
Efs
|
embryonal Fyn-associated substrate |
| chr1_-_155108455 | 0.34 |
ENSMUST00000035914.5
|
BC034090
|
cDNA sequence BC034090 |
| chr1_+_177272297 | 0.34 |
ENSMUST00000193440.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr8_-_114575247 | 0.34 |
ENSMUST00000093113.5
|
Adamts18
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 18 |
| chr6_-_142268667 | 0.33 |
ENSMUST00000128446.2
|
Slco1a5
|
solute carrier organic anion transporter family, member 1a5 |
| chr7_+_101619327 | 0.33 |
ENSMUST00000210475.2
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr2_-_85916198 | 0.32 |
ENSMUST00000099911.5
|
Olfr1037
|
olfactory receptor 1037 |
| chr4_-_139560831 | 0.31 |
ENSMUST00000030508.14
|
Pax7
|
paired box 7 |
| chr15_+_11000802 | 0.31 |
ENSMUST00000117100.4
|
Slc45a2
|
solute carrier family 45, member 2 |
| chr7_+_13132040 | 0.31 |
ENSMUST00000005791.14
|
Cabp5
|
calcium binding protein 5 |
| chr1_-_13730732 | 0.30 |
ENSMUST00000027071.7
|
Lactb2
|
lactamase, beta 2 |
| chrX_+_20291927 | 0.30 |
ENSMUST00000115384.9
|
Jade3
|
jade family PHD finger 3 |
| chr6_+_115911302 | 0.30 |
ENSMUST00000203877.2
|
Rho
|
rhodopsin |
| chr9_+_38399327 | 0.30 |
ENSMUST00000045493.2
|
Olfr906
|
olfactory receptor 906 |
| chr10_-_22025169 | 0.30 |
ENSMUST00000179054.8
ENSMUST00000069372.7 |
E030030I06Rik
|
RIKEN cDNA E030030I06 gene |
| chr8_-_96033213 | 0.29 |
ENSMUST00000119870.9
ENSMUST00000093268.5 |
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr7_+_126575510 | 0.28 |
ENSMUST00000206780.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr1_-_4430481 | 0.28 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr4_+_101407608 | 0.28 |
ENSMUST00000094953.11
ENSMUST00000106933.2 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr11_+_23206001 | 0.28 |
ENSMUST00000020538.13
ENSMUST00000109551.8 ENSMUST00000102870.8 ENSMUST00000102869.8 |
Xpo1
|
exportin 1 |
| chr7_-_127324788 | 0.25 |
ENSMUST00000076091.4
|
Ctf2
|
cardiotrophin 2 |
| chr15_-_65848649 | 0.25 |
ENSMUST00000239034.2
ENSMUST00000100584.3 |
Hhla1
|
HERV-H LTR-associating 1 |
| chr14_-_118289557 | 0.25 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr7_-_4517608 | 0.25 |
ENSMUST00000166959.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr4_+_19280850 | 0.25 |
ENSMUST00000102999.2
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr18_-_78222349 | 0.24 |
ENSMUST00000237290.2
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr13_-_113755082 | 0.22 |
ENSMUST00000109241.5
|
Snx18
|
sorting nexin 18 |
| chr11_-_5049036 | 0.21 |
ENSMUST00000102930.10
ENSMUST00000093365.12 ENSMUST00000073308.11 |
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr7_-_105401398 | 0.21 |
ENSMUST00000033184.6
|
Tpp1
|
tripeptidyl peptidase I |
| chr19_-_47680528 | 0.21 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr5_-_28442871 | 0.21 |
ENSMUST00000120068.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr13_-_105430932 | 0.21 |
ENSMUST00000224662.2
|
Rnf180
|
ring finger protein 180 |
| chr11_-_5049223 | 0.21 |
ENSMUST00000079949.13
|
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr17_-_74602469 | 0.21 |
ENSMUST00000233144.2
|
Memo1
|
mediator of cell motility 1 |
| chr11_+_69886652 | 0.20 |
ENSMUST00000101526.9
|
Phf23
|
PHD finger protein 23 |
| chr10_+_23952398 | 0.18 |
ENSMUST00000051133.6
|
Taar8a
|
trace amine-associated receptor 8A |
| chr13_-_105430889 | 0.17 |
ENSMUST00000226044.2
|
Rnf180
|
ring finger protein 180 |
| chr7_+_103684652 | 0.17 |
ENSMUST00000213214.2
|
Olfr641
|
olfactory receptor 641 |
| chr7_-_4517559 | 0.17 |
ENSMUST00000163538.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr2_+_89642395 | 0.17 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr12_-_115706126 | 0.16 |
ENSMUST00000166645.2
ENSMUST00000196690.2 |
Ighv1-71
|
immunoglobulin heavy variable 1-71 |
| chr2_+_85600147 | 0.16 |
ENSMUST00000065626.3
|
Olfr1013
|
olfactory receptor 1013 |
| chr6_+_38086190 | 0.15 |
ENSMUST00000031851.5
|
Tmem213
|
transmembrane protein 213 |
| chr7_+_63803583 | 0.15 |
ENSMUST00000085222.12
ENSMUST00000206277.2 |
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr11_+_69886603 | 0.15 |
ENSMUST00000018716.10
|
Phf23
|
PHD finger protein 23 |
| chr10_+_21869776 | 0.15 |
ENSMUST00000092673.11
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr2_-_73316809 | 0.14 |
ENSMUST00000141264.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr10_+_128061699 | 0.14 |
ENSMUST00000026455.8
|
Mip
|
major intrinsic protein of lens fiber |
| chr2_+_91095597 | 0.13 |
ENSMUST00000028691.7
|
Arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr7_-_104246386 | 0.13 |
ENSMUST00000057385.5
|
Olfr655
|
olfactory receptor 655 |
| chr1_+_82817794 | 0.12 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr10_+_18283405 | 0.11 |
ENSMUST00000037341.14
|
Nhsl1
|
NHS-like 1 |
| chr3_-_144804784 | 0.11 |
ENSMUST00000040465.11
ENSMUST00000198993.2 |
Clca2
|
chloride channel accessory 2 |
| chr6_+_42873387 | 0.11 |
ENSMUST00000058668.4
|
Olfr448
|
olfactory receptor 448 |
| chrX_+_99615836 | 0.11 |
ENSMUST00000096361.5
|
Awat1
|
acyl-CoA wax alcohol acyltransferase 1 |
| chr2_-_120916316 | 0.11 |
ENSMUST00000028721.8
|
Tgm5
|
transglutaminase 5 |
| chr18_+_46730765 | 0.10 |
ENSMUST00000238168.2
ENSMUST00000078079.11 ENSMUST00000168382.2 ENSMUST00000235849.2 ENSMUST00000235973.2 ENSMUST00000235455.2 ENSMUST00000237478.2 |
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr2_-_51863203 | 0.10 |
ENSMUST00000028314.9
|
Nmi
|
N-myc (and STAT) interactor |
| chr7_+_43418404 | 0.10 |
ENSMUST00000014063.6
|
Klk12
|
kallikrein related-peptidase 12 |
| chr9_-_108443916 | 0.09 |
ENSMUST00000194381.2
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr1_+_178015287 | 0.09 |
ENSMUST00000159284.2
|
Desi2
|
desumoylating isopeptidase 2 |
| chr3_+_66127330 | 0.09 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chrX_-_42256694 | 0.09 |
ENSMUST00000115058.8
ENSMUST00000115059.8 |
Tenm1
|
teneurin transmembrane protein 1 |
| chr8_+_79236051 | 0.09 |
ENSMUST00000209992.2
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr7_-_103214484 | 0.08 |
ENSMUST00000106886.2
|
Olfr616
|
olfactory receptor 616 |
| chr1_-_154692678 | 0.08 |
ENSMUST00000238369.2
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr5_-_66161621 | 0.08 |
ENSMUST00000201351.4
|
9130230L23Rik
|
RIKEN cDNA 9130230L23 gene |
| chr7_+_43418321 | 0.07 |
ENSMUST00000107970.8
|
Klk12
|
kallikrein related-peptidase 12 |
| chr18_-_60860594 | 0.07 |
ENSMUST00000235795.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr7_+_63803663 | 0.07 |
ENSMUST00000206314.2
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr10_-_129023288 | 0.06 |
ENSMUST00000072976.4
|
Olfr773
|
olfactory receptor 773 |
| chrM_+_8603 | 0.06 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr17_-_72059865 | 0.05 |
ENSMUST00000057405.9
|
Pcare
|
photoreceptor cilium actin regulator |
| chr19_+_36061096 | 0.04 |
ENSMUST00000025714.9
|
Rpp30
|
ribonuclease P/MRP 30 subunit |
| chr14_-_55762395 | 0.03 |
ENSMUST00000228287.2
|
Nrl
|
neural retina leucine zipper gene |
| chr6_-_59403264 | 0.03 |
ENSMUST00000051065.6
|
Gprin3
|
GPRIN family member 3 |
| chr16_+_17051423 | 0.02 |
ENSMUST00000090190.14
ENSMUST00000232082.2 ENSMUST00000232426.2 |
Hic2
Gm49573
|
hypermethylated in cancer 2 predicted gene, 49573 |
| chr7_-_80053063 | 0.01 |
ENSMUST00000147150.2
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr12_-_115206715 | 0.00 |
ENSMUST00000103527.2
ENSMUST00000194071.2 |
Ighv1-56
|
immunoglobulin heavy variable 1-56 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.8 | 4.1 | GO:0002358 | pre-B cell allelic exclusion(GO:0002331) B cell homeostatic proliferation(GO:0002358) |
| 0.6 | 2.5 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.6 | 6.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.6 | 2.2 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.5 | 1.6 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.5 | 1.8 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.4 | 1.4 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.3 | 0.8 | GO:0034147 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.3 | 8.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.3 | 1.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 0.8 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.2 | 0.8 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.2 | 0.5 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 | 0.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 1.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 2.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.5 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 0.3 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.4 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.7 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 1.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 1.2 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.4 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 1.7 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 1.7 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.4 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.3 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.7 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.4 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.1 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.2 | 1.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 0.8 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 8.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 6.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 2.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.7 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.2 | GO:0030315 | T-tubule(GO:0030315) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.3 | 1.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 1.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 1.4 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 1.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 1.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 0.6 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 1.0 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 2.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 7.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 4.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.3 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 1.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.3 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 1.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 6.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 2.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.9 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0015250 | water transmembrane transporter activity(GO:0005372) water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 2.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 4.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 6.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 4.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 2.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.1 | 0.9 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 4.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |