Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
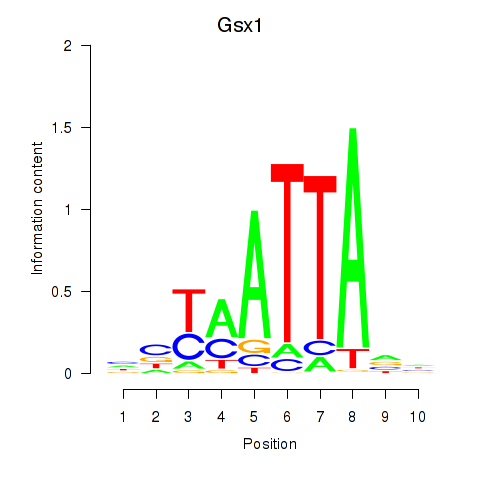
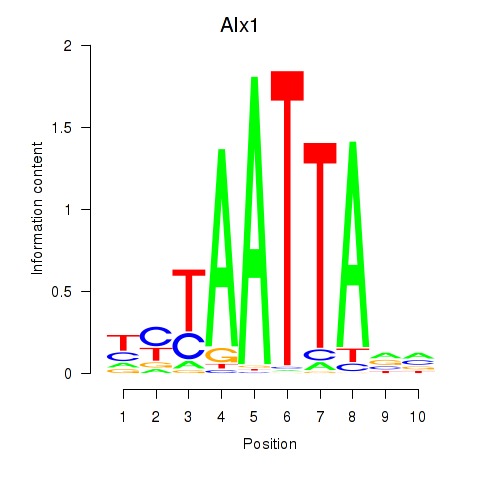
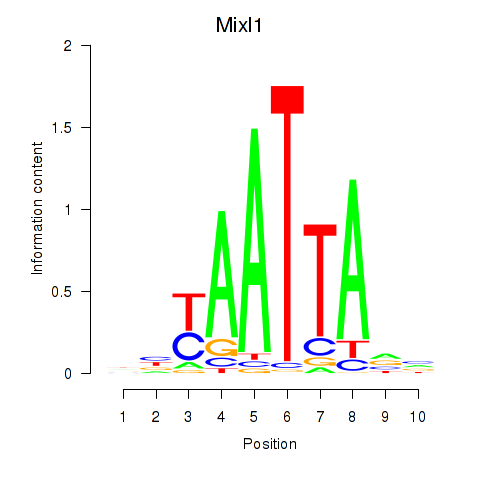
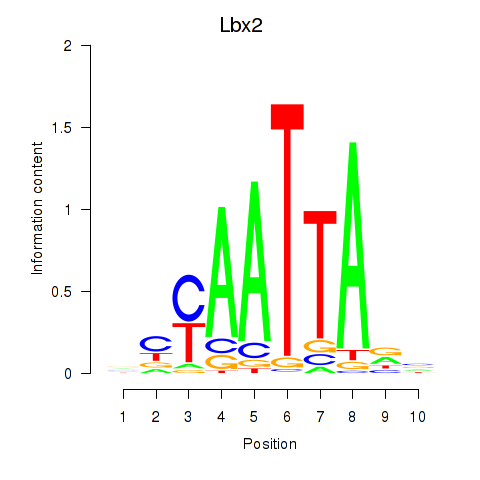
Results for Gsx1_Alx1_Mixl1_Lbx2
Z-value: 1.01
Transcription factors associated with Gsx1_Alx1_Mixl1_Lbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gsx1
|
ENSMUSG00000053129.6 | Gsx1 |
|
Alx1
|
ENSMUSG00000036602.15 | Alx1 |
|
Mixl1
|
ENSMUSG00000026497.8 | Mixl1 |
|
Lbx2
|
ENSMUSG00000034968.4 | Lbx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gsx1 | mm39_v1_chr5_+_147125488_147125506 | -0.59 | 6.3e-08 | Click! |
| Alx1 | mm39_v1_chr10_-_102866076_102866205 | -0.25 | 3.2e-02 | Click! |
| Mixl1 | mm39_v1_chr1_-_180524587_180524599 | 0.18 | 1.4e-01 | Click! |
| Lbx2 | mm39_v1_chr6_+_83063348_83063348 | 0.03 | 7.7e-01 | Click! |
Activity profile of Gsx1_Alx1_Mixl1_Lbx2 motif
Sorted Z-values of Gsx1_Alx1_Mixl1_Lbx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gsx1_Alx1_Mixl1_Lbx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_129449739 | 10.94 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr3_-_88317601 | 10.71 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr5_-_62923463 | 10.06 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chrX_+_149330371 | 9.90 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr11_-_99482165 | 9.27 |
ENSMUST00000104930.2
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr2_-_72817060 | 8.11 |
ENSMUST00000112062.2
|
Gm11084
|
predicted gene 11084 |
| chr10_-_107321938 | 7.69 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr11_+_11634967 | 7.62 |
ENSMUST00000141436.8
ENSMUST00000126058.8 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr14_+_53562089 | 6.96 |
ENSMUST00000178100.3
|
Trav7n-6
|
T cell receptor alpha variable 7N-6 |
| chr5_+_96104775 | 6.90 |
ENSMUST00000023840.7
|
Cxcl13
|
chemokine (C-X-C motif) ligand 13 |
| chr19_-_46033353 | 6.62 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr15_-_66985760 | 6.55 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chrX_+_56257374 | 6.53 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr3_-_49711706 | 6.48 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr14_+_79753055 | 6.34 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr11_-_99441687 | 6.24 |
ENSMUST00000092700.5
|
Krtap3-3
|
keratin associated protein 3-3 |
| chrX_+_41241049 | 6.24 |
ENSMUST00000128799.3
|
Stag2
|
stromal antigen 2 |
| chr16_+_33504740 | 6.22 |
ENSMUST00000232568.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr1_-_171854818 | 6.06 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr11_-_99265721 | 6.00 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr3_-_49711765 | 5.75 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr4_+_98919183 | 5.75 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr8_-_62576140 | 5.68 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr15_+_6673167 | 5.58 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein |
| chr8_-_85389470 | 5.56 |
ENSMUST00000060427.6
|
Ier2
|
immediate early response 2 |
| chr15_+_55171138 | 5.19 |
ENSMUST00000023053.12
ENSMUST00000110217.10 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr10_+_23727325 | 5.18 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr6_-_50631418 | 5.04 |
ENSMUST00000031853.8
|
Npvf
|
neuropeptide VF precursor |
| chr17_+_34524841 | 5.01 |
ENSMUST00000235530.2
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr11_+_67689094 | 4.97 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr9_-_71070506 | 4.95 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chrX_+_132751729 | 4.85 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chrX_+_139857640 | 4.83 |
ENSMUST00000112971.2
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr7_+_63835285 | 4.79 |
ENSMUST00000206263.2
ENSMUST00000206107.2 ENSMUST00000205731.2 ENSMUST00000206706.2 ENSMUST00000205690.2 |
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr2_-_27365633 | 4.67 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr16_+_35861554 | 4.57 |
ENSMUST00000042203.10
|
Wdr5b
|
WD repeat domain 5B |
| chrX_+_139857688 | 4.51 |
ENSMUST00000239541.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr17_+_71326510 | 4.48 |
ENSMUST00000073211.13
ENSMUST00000024847.14 |
Myom1
|
myomesin 1 |
| chr12_-_11258973 | 4.47 |
ENSMUST00000049877.3
|
Msgn1
|
mesogenin 1 |
| chr6_+_41511248 | 4.44 |
ENSMUST00000192366.2
ENSMUST00000103286.2 |
Trbj1-3
|
T cell receptor beta joining 1-3 |
| chr1_-_69726384 | 4.35 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr19_-_11243530 | 4.29 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr11_+_67090878 | 4.26 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr11_-_99494134 | 4.24 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr8_-_62355690 | 4.20 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr11_+_60428788 | 4.14 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr3_-_75177378 | 4.13 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr16_-_48592372 | 3.99 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr17_+_71326542 | 3.98 |
ENSMUST00000179759.3
|
Myom1
|
myomesin 1 |
| chr15_+_31224616 | 3.90 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr3_+_82915031 | 3.87 |
ENSMUST00000048486.13
ENSMUST00000194175.2 |
Fgg
|
fibrinogen gamma chain |
| chr3_-_144514386 | 3.80 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr6_-_83654789 | 3.80 |
ENSMUST00000037882.8
|
Cd207
|
CD207 antigen |
| chr17_-_84154196 | 3.70 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr3_-_92441809 | 3.69 |
ENSMUST00000193521.2
|
2310046K23Rik
|
RIKEN cDNA 2310046K23 gene |
| chr6_-_115569504 | 3.68 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr14_+_33662976 | 3.67 |
ENSMUST00000100720.2
|
Gdf2
|
growth differentiation factor 2 |
| chr5_-_21087023 | 3.67 |
ENSMUST00000118174.8
|
Phtf2
|
putative homeodomain transcription factor 2 |
| chr14_+_54183465 | 3.67 |
ENSMUST00000197130.5
ENSMUST00000103677.3 |
Trdv2-1
|
T cell receptor delta variable 2-1 |
| chr2_-_84255602 | 3.59 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr9_+_43222104 | 3.54 |
ENSMUST00000034511.7
|
Trim29
|
tripartite motif-containing 29 |
| chr4_-_129452180 | 3.53 |
ENSMUST00000067240.11
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr18_+_36414122 | 3.49 |
ENSMUST00000051301.6
|
Pura
|
purine rich element binding protein A |
| chr4_-_129452148 | 3.48 |
ENSMUST00000167288.8
ENSMUST00000134336.3 |
Lck
|
lymphocyte protein tyrosine kinase |
| chr14_+_54436247 | 3.45 |
ENSMUST00000103720.2
|
Traj21
|
T cell receptor alpha joining 21 |
| chr9_+_96140781 | 3.42 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr3_-_105940130 | 3.37 |
ENSMUST00000200146.2
|
Chil5
|
chitinase-like 5 |
| chr6_+_125529911 | 3.36 |
ENSMUST00000112254.8
ENSMUST00000112253.6 |
Vwf
|
Von Willebrand factor |
| chr16_+_33504908 | 3.35 |
ENSMUST00000126532.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chrM_+_7006 | 3.34 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chrX_-_101200670 | 3.32 |
ENSMUST00000056904.3
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 like |
| chr11_-_107228382 | 3.31 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr11_+_23256883 | 3.28 |
ENSMUST00000180046.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr6_-_41752111 | 3.27 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr10_-_128361731 | 3.26 |
ENSMUST00000026427.8
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr14_-_86986541 | 3.24 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr1_-_45542442 | 3.24 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr3_+_59989282 | 3.20 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr14_+_53954133 | 3.14 |
ENSMUST00000103641.6
|
Trav7-6
|
T cell receptor alpha variable 7-6 |
| chr11_-_99501015 | 3.12 |
ENSMUST00000076478.2
|
Gm11937
|
predicted gene 11937 |
| chr19_-_24178000 | 3.08 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr1_-_4430481 | 3.07 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr17_+_34524884 | 2.99 |
ENSMUST00000074557.11
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr18_+_4920513 | 2.97 |
ENSMUST00000126977.8
|
Svil
|
supervillin |
| chr3_+_84573499 | 2.97 |
ENSMUST00000107682.2
|
Tmem154
|
transmembrane protein 154 |
| chr18_+_84869456 | 2.94 |
ENSMUST00000160180.9
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr4_-_43710231 | 2.93 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr7_+_63835154 | 2.90 |
ENSMUST00000177102.8
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr6_-_87510200 | 2.89 |
ENSMUST00000113637.9
ENSMUST00000071024.7 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr6_-_138056914 | 2.87 |
ENSMUST00000171804.4
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr9_+_96140750 | 2.86 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr2_+_174292471 | 2.86 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr6_-_128252540 | 2.85 |
ENSMUST00000130454.8
|
Tead4
|
TEA domain family member 4 |
| chr4_+_135870808 | 2.84 |
ENSMUST00000008016.3
|
Id3
|
inhibitor of DNA binding 3 |
| chr16_-_48592319 | 2.80 |
ENSMUST00000239408.2
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr18_-_43610829 | 2.78 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr4_-_45532470 | 2.66 |
ENSMUST00000147448.2
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr11_-_69786324 | 2.65 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr8_+_106786190 | 2.65 |
ENSMUST00000109308.3
|
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr8_-_3675274 | 2.59 |
ENSMUST00000004749.7
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr17_-_37523969 | 2.56 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr4_+_114914880 | 2.53 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr11_+_23256909 | 2.52 |
ENSMUST00000137823.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr7_-_4909515 | 2.50 |
ENSMUST00000210663.2
|
Gm36210
|
predicted gene, 36210 |
| chr10_-_44024843 | 2.45 |
ENSMUST00000200401.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr17_-_24054717 | 2.45 |
ENSMUST00000059906.8
|
Prss33
|
protease, serine 33 |
| chr14_+_52892115 | 2.44 |
ENSMUST00000198019.2
|
Trav7-1
|
T cell receptor alpha variable 7-1 |
| chr5_-_137856280 | 2.43 |
ENSMUST00000110978.7
ENSMUST00000199387.2 ENSMUST00000196195.2 |
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr6_+_123239076 | 2.43 |
ENSMUST00000032240.4
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr1_+_53100796 | 2.37 |
ENSMUST00000027269.7
ENSMUST00000191197.2 |
Mstn
|
myostatin |
| chr2_+_152596075 | 2.35 |
ENSMUST00000010020.12
|
Cox4i2
|
cytochrome c oxidase subunit 4I2 |
| chr2_-_111843053 | 2.32 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr5_-_65855511 | 2.32 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr15_+_101371353 | 2.30 |
ENSMUST00000088049.5
|
Krt86
|
keratin 86 |
| chr1_+_40554513 | 2.30 |
ENSMUST00000027237.12
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr16_-_19079594 | 2.27 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr7_-_83304698 | 2.25 |
ENSMUST00000145610.8
|
Il16
|
interleukin 16 |
| chr6_+_37847721 | 2.24 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr2_-_153079828 | 2.23 |
ENSMUST00000109795.2
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr6_-_16898440 | 2.13 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chr6_+_129374441 | 2.11 |
ENSMUST00000112081.9
ENSMUST00000112079.3 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr15_-_66684442 | 2.11 |
ENSMUST00000100572.10
|
Sla
|
src-like adaptor |
| chr15_-_101801351 | 2.10 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr13_-_113237505 | 2.06 |
ENSMUST00000224282.2
ENSMUST00000023897.7 |
Gzma
|
granzyme A |
| chr17_+_34811217 | 2.05 |
ENSMUST00000038149.13
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr3_-_130524024 | 2.04 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr2_+_87725306 | 2.03 |
ENSMUST00000217436.2
|
Olfr1153
|
olfactory receptor 1153 |
| chr14_+_73475335 | 2.03 |
ENSMUST00000044405.8
|
Lpar6
|
lysophosphatidic acid receptor 6 |
| chr15_+_80507671 | 2.02 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr1_-_184543367 | 2.01 |
ENSMUST00000048462.13
ENSMUST00000110992.9 |
Mtarc1
|
mitochondrial amidoxime reducing component 1 |
| chr16_+_43574389 | 2.00 |
ENSMUST00000229953.2
|
Drd3
|
dopamine receptor D3 |
| chr6_-_51443602 | 2.00 |
ENSMUST00000203253.2
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr9_+_108216233 | 2.00 |
ENSMUST00000082429.8
|
Gpx1
|
glutathione peroxidase 1 |
| chr9_-_110775143 | 1.98 |
ENSMUST00000199782.2
ENSMUST00000035075.13 |
Tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr5_+_13448647 | 1.96 |
ENSMUST00000125629.8
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_-_29226700 | 1.95 |
ENSMUST00000233441.2
|
Stk38
|
serine/threonine kinase 38 |
| chr6_-_68609426 | 1.93 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr7_-_10488291 | 1.91 |
ENSMUST00000226874.2
ENSMUST00000227003.2 ENSMUST00000228561.2 ENSMUST00000228248.2 ENSMUST00000228526.2 ENSMUST00000228098.2 ENSMUST00000227940.2 ENSMUST00000228374.2 ENSMUST00000227702.2 |
Vmn1r71
|
vomeronasal 1 receptor 71 |
| chr3_+_32490300 | 1.91 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr3_+_57332735 | 1.90 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr9_+_108216466 | 1.90 |
ENSMUST00000193987.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr16_-_26808724 | 1.89 |
ENSMUST00000089832.6
|
Gmnc
|
geminin coiled-coil domain containing |
| chr17_-_84154173 | 1.88 |
ENSMUST00000000687.9
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr10_-_39039790 | 1.86 |
ENSMUST00000076713.6
|
Ccn6
|
cellular communication network factor 6 |
| chr10_+_127256736 | 1.86 |
ENSMUST00000064793.13
|
R3hdm2
|
R3H domain containing 2 |
| chr9_+_118307412 | 1.84 |
ENSMUST00000035020.15
|
Eomes
|
eomesodermin |
| chr15_-_65784103 | 1.81 |
ENSMUST00000079776.14
|
Oc90
|
otoconin 90 |
| chr3_-_106697459 | 1.77 |
ENSMUST00000038845.10
|
Cd53
|
CD53 antigen |
| chr1_+_150195158 | 1.76 |
ENSMUST00000165062.8
ENSMUST00000191228.7 ENSMUST00000186572.7 ENSMUST00000185698.2 |
Pdc
|
phosducin |
| chr19_+_25649767 | 1.74 |
ENSMUST00000053068.7
|
Dmrt2
|
doublesex and mab-3 related transcription factor 2 |
| chr9_-_90152759 | 1.74 |
ENSMUST00000041767.14
ENSMUST00000128874.3 |
Tbc1d2b
|
TBC1 domain family, member 2B |
| chr10_-_88518878 | 1.73 |
ENSMUST00000004473.15
|
Spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr2_-_168607166 | 1.73 |
ENSMUST00000137536.2
|
Sall4
|
spalt like transcription factor 4 |
| chr6_+_41025217 | 1.71 |
ENSMUST00000103264.3
|
Trbv3
|
T cell receptor beta, variable 3 |
| chr14_+_32507920 | 1.70 |
ENSMUST00000039191.8
ENSMUST00000227060.2 ENSMUST00000228481.2 |
Tmem273
|
transmembrane protein 273 |
| chr11_+_67061837 | 1.70 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr9_+_65797519 | 1.70 |
ENSMUST00000045802.7
|
Pclaf
|
PCNA clamp associated factor |
| chr15_+_31224460 | 1.69 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr19_-_7943365 | 1.67 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr5_-_36641456 | 1.67 |
ENSMUST00000119916.2
ENSMUST00000031097.8 |
Tada2b
|
transcriptional adaptor 2B |
| chr9_+_108216433 | 1.66 |
ENSMUST00000191997.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr5_-_87847268 | 1.65 |
ENSMUST00000196869.5
ENSMUST00000199624.5 ENSMUST00000198057.5 ENSMUST00000082370.10 |
Csn2
|
casein beta |
| chr7_-_83384711 | 1.64 |
ENSMUST00000001792.12
|
Il16
|
interleukin 16 |
| chr16_-_89368059 | 1.63 |
ENSMUST00000171542.2
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr11_-_99511257 | 1.62 |
ENSMUST00000073853.3
|
Gm11562
|
predicted gene 11562 |
| chr6_+_70648743 | 1.62 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4 |
| chr17_-_37511885 | 1.62 |
ENSMUST00000222190.2
|
Olfr94
|
olfactory receptor 94 |
| chr7_+_49559859 | 1.60 |
ENSMUST00000056442.12
|
Slc6a5
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
| chr14_+_53698556 | 1.60 |
ENSMUST00000181728.3
|
Trav7-4
|
T cell receptor alpha variable 7-4 |
| chr11_+_67061908 | 1.60 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr17_-_48758538 | 1.58 |
ENSMUST00000024794.12
|
Tspo2
|
translocator protein 2 |
| chr15_-_65784246 | 1.57 |
ENSMUST00000060522.11
|
Oc90
|
otoconin 90 |
| chr6_-_68681962 | 1.57 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chrX_-_163041185 | 1.57 |
ENSMUST00000112265.9
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr12_+_117807224 | 1.55 |
ENSMUST00000021592.16
|
Cdca7l
|
cell division cycle associated 7 like |
| chr13_-_43634695 | 1.55 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr16_+_57173456 | 1.54 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr5_-_137834470 | 1.54 |
ENSMUST00000110980.2
ENSMUST00000058897.11 ENSMUST00000199028.2 |
Pilra
|
paired immunoglobin-like type 2 receptor alpha |
| chr3_+_145827410 | 1.54 |
ENSMUST00000039450.5
|
Mcoln3
|
mucolipin 3 |
| chr15_-_42540363 | 1.52 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr17_+_46471950 | 1.51 |
ENSMUST00000024748.14
ENSMUST00000172170.8 |
Gtpbp2
|
GTP binding protein 2 |
| chr10_+_50770836 | 1.51 |
ENSMUST00000219436.2
|
Sim1
|
single-minded family bHLH transcription factor 1 |
| chr3_-_15902583 | 1.51 |
ENSMUST00000108354.8
ENSMUST00000108349.2 ENSMUST00000108352.9 ENSMUST00000108350.8 ENSMUST00000050623.11 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr15_+_102314809 | 1.46 |
ENSMUST00000001326.7
|
Sp1
|
trans-acting transcription factor 1 |
| chr12_-_114710326 | 1.45 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr12_-_114547622 | 1.45 |
ENSMUST00000193893.6
ENSMUST00000103498.3 |
Ighv1-9
|
immunoglobulin heavy variable V1-9 |
| chr7_-_45480200 | 1.44 |
ENSMUST00000107723.9
ENSMUST00000131384.3 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr12_-_104439589 | 1.43 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr3_-_132940647 | 1.43 |
ENSMUST00000147041.10
ENSMUST00000161022.9 |
Arhgef38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr5_-_137529465 | 1.42 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr5_+_88603847 | 1.42 |
ENSMUST00000198265.5
ENSMUST00000031226.9 |
Ambn
|
ameloblastin |
| chr6_+_68916540 | 1.42 |
ENSMUST00000103339.2
|
Igkv13-84
|
immunoglobulin kappa chain variable 13-84 |
| chr3_+_63203516 | 1.40 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chr1_+_82817794 | 1.39 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr15_+_25774070 | 1.39 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr8_-_58106057 | 1.38 |
ENSMUST00000034021.12
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr11_+_116734104 | 1.37 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr4_-_14621669 | 1.37 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr4_+_80752535 | 1.36 |
ENSMUST00000102831.8
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr7_+_86109129 | 1.36 |
ENSMUST00000217253.2
|
Olfr299
|
olfactory receptor 299 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:0003017 | lymph circulation(GO:0003017) |
| 2.5 | 7.6 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 2.2 | 6.6 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 2.2 | 6.5 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 2.0 | 6.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 1.7 | 6.9 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 1.4 | 5.6 | GO:0010269 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 1.3 | 9.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 1.3 | 5.1 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.1 | 6.8 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.0 | 7.7 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 1.0 | 5.7 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.9 | 3.6 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.8 | 2.5 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.8 | 2.5 | GO:1904826 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.8 | 3.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.8 | 2.4 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.8 | 2.4 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.8 | 3.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.8 | 6.9 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.7 | 2.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.7 | 2.2 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.7 | 2.2 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.7 | 4.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.7 | 10.1 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.7 | 2.0 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.7 | 2.0 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.6 | 6.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.6 | 2.4 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.6 | 4.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.6 | 2.8 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.5 | 7.0 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.5 | 1.6 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.5 | 8.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.4 | 1.8 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.4 | 3.5 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.4 | 1.7 | GO:0061055 | myotome development(GO:0061055) |
| 0.4 | 3.5 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.4 | 10.8 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.4 | 3.8 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 | 1.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.4 | 1.6 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.4 | 2.8 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.4 | 1.6 | GO:0036233 | glycine import(GO:0036233) |
| 0.4 | 1.2 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 0.4 | 1.2 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.4 | 8.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.4 | 1.5 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.3 | 1.0 | GO:0060618 | nipple development(GO:0060618) |
| 0.3 | 0.9 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.3 | 1.2 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.3 | 1.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.3 | 3.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 0.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.3 | 2.6 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.3 | 0.9 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.3 | 1.1 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.3 | 10.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 | 0.8 | GO:1903000 | positive regulation of low-density lipoprotein particle receptor catabolic process(GO:0032805) regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.3 | 3.9 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.3 | 2.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.3 | 5.8 | GO:0007379 | segment specification(GO:0007379) |
| 0.3 | 0.5 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.3 | 1.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.3 | 0.8 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.3 | 0.8 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.2 | 0.7 | GO:0061360 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.2 | 2.6 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 0.7 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.2 | 1.4 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.2 | 0.8 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.2 | 0.8 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.2 | 2.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.2 | 1.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 1.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 1.0 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.2 | 4.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 0.6 | GO:1901874 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.2 | 0.9 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 3.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 0.4 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.2 | 1.2 | GO:1901509 | branch elongation involved in ureteric bud branching(GO:0060681) regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 0.5 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.2 | 0.7 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.2 | 0.5 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.2 | 2.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 1.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 0.7 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.2 | 0.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 6.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.2 | 0.5 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.2 | 0.6 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 0.5 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.2 | 3.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 | 2.7 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 1.0 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 1.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 5.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.9 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 0.9 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 1.3 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 0.5 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 | 1.0 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 5.2 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 1.0 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.9 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 7.5 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 1.2 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.1 | 0.7 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 3.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 1.0 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 7.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.5 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.1 | 0.3 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.7 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 1.7 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 1.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.9 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 2.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 1.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 2.0 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.1 | 4.3 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 1.2 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 0.8 | GO:1903140 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.1 | 2.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 1.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 2.8 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 0.7 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 2.6 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 3.0 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.6 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.6 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 1.6 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.9 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 5.2 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 0.5 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.6 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.9 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 1.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.6 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 1.4 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.3 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 3.8 | GO:0043276 | anoikis(GO:0043276) |
| 0.1 | 0.8 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.4 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 3.0 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.1 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 1.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.6 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.5 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.4 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.9 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 2.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 1.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.3 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.2 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 1.0 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 1.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 1.5 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 2.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.3 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.1 | 0.6 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 5.8 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.1 | 0.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 1.6 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 2.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.9 | GO:0033617 | protein import into mitochondrial matrix(GO:0030150) mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 1.8 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.2 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.9 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 1.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 1.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 2.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 3.6 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.0 | 0.4 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 1.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 1.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 6.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 10.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.7 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.8 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.4 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 3.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.9 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.5 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.4 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.9 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 1.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 2.2 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 1.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.8 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 3.1 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 3.4 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 4.8 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.6 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 1.8 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 31.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.4 | GO:0046512 | sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.3 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 0.7 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 2.1 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 8.1 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 3.8 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.5 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 0.5 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 1.6 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 1.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.7 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 3.6 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.6 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.0 | GO:0048144 | fibroblast proliferation(GO:0048144) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.3 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.8 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.3 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.1 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.4 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.7 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 2.0 | 6.1 | GO:0060187 | cell pole(GO:0060187) |
| 1.7 | 5.0 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 1.5 | 7.7 | GO:0035841 | new growing cell tip(GO:0035841) |
| 1.1 | 3.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 2.5 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.7 | 2.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.7 | 5.6 | GO:0097413 | Lewy body(GO:0097413) |
| 0.7 | 8.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.6 | 16.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.4 | 3.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 7.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.3 | 3.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 5.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 1.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 1.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.3 | 2.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.3 | 2.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.3 | 0.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.3 | 3.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 0.9 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.2 | 4.0 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 6.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 1.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.2 | 6.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 3.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 6.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 0.6 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.2 | 0.8 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 2.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 2.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.7 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 1.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 1.0 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 2.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.9 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.5 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.9 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 1.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 3.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.5 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 4.2 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 6.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 26.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 1.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 11.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 1.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 1.4 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 2.3 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 4.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 4.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 6.2 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 11.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 2.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 1.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 29.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 4.1 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 3.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 2.2 | 6.5 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 1.7 | 6.9 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.2 | 6.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.2 | 3.6 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 1.0 | 5.1 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.9 | 3.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.7 | 6.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.7 | 2.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.7 | 4.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.7 | 2.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.5 | 2.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.5 | 5.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.5 | 8.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.4 | 3.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.4 | 3.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.4 | 1.6 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.4 | 2.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.4 | 7.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 2.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.3 | 1.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.3 | 2.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.3 | 2.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.3 | 2.8 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 1.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.3 | 2.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 2.0 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.3 | 0.8 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.3 | 3.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 6.8 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.3 | 2.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 2.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 1.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.2 | 5.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 5.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 2.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 6.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 3.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 0.6 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.2 | 1.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 7.0 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.2 | 1.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 2.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 4.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 3.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.2 | 4.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 0.9 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 2.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.2 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.5 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.2 | 2.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.2 | 4.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 1.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 5.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 0.9 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 16.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.0 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 0.4 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 10.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 2.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.8 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 4.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 2.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 2.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 2.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 6.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 2.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.3 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.1 | 0.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 1.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.6 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 5.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 2.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.6 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 1.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.3 | GO:0003681 | bent DNA binding(GO:0003681) |
| 0.1 | 9.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 2.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 47.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.4 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 1.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 2.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 2.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.7 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.1 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 0.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 3.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.8 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 1.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.0 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 5.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.2 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 1.7 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 3.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.5 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 2.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 4.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 6.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.1 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 2.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 4.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 32.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 4.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.0 | 0.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 3.3 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 2.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 9.3 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.0 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 2.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.0 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.9 | GO:0008238 | exopeptidase activity(GO:0008238) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 9.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 12.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 9.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 10.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 11.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 8.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 6.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 7.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 3.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 3.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 6.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 8.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 11.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 5.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 1.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 4.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 10.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 0.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 5.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 10.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 3.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 7.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 6.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 9.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.4 | 5.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 7.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.3 | 5.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 9.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.3 | 7.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 6.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 0.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 23.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 10.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 2.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 6.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 6.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 7.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.8 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 0.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 7.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 2.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 2.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 1.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 8.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 2.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 1.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 3.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.0 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |