Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Gsx2_Hoxd3_Vax1
Z-value: 0.77
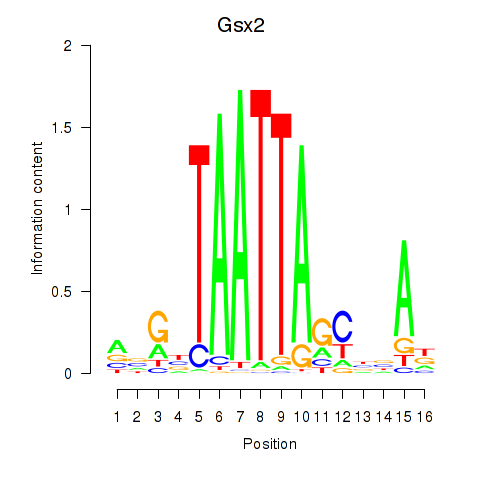
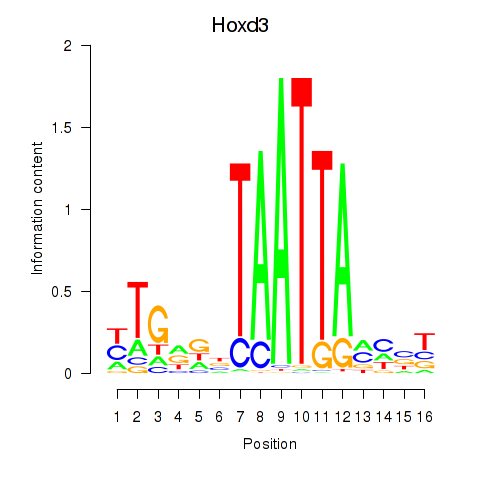
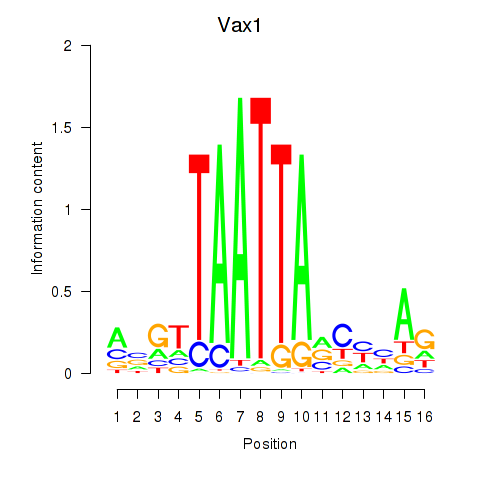
Transcription factors associated with Gsx2_Hoxd3_Vax1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gsx2
|
ENSMUSG00000035946.8 | Gsx2 |
|
Hoxd3
|
ENSMUSG00000079277.10 | Hoxd3 |
|
Vax1
|
ENSMUSG00000006270.8 | Vax1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd3 | mm39_v1_chr2_+_74566740_74566859 | 0.38 | 1.0e-03 | Click! |
| Vax1 | mm39_v1_chr19_-_59158488_59158488 | 0.33 | 4.8e-03 | Click! |
| Gsx2 | mm39_v1_chr5_+_75236250_75236263 | 0.18 | 1.3e-01 | Click! |
Activity profile of Gsx2_Hoxd3_Vax1 motif
Sorted Z-values of Gsx2_Hoxd3_Vax1 motif
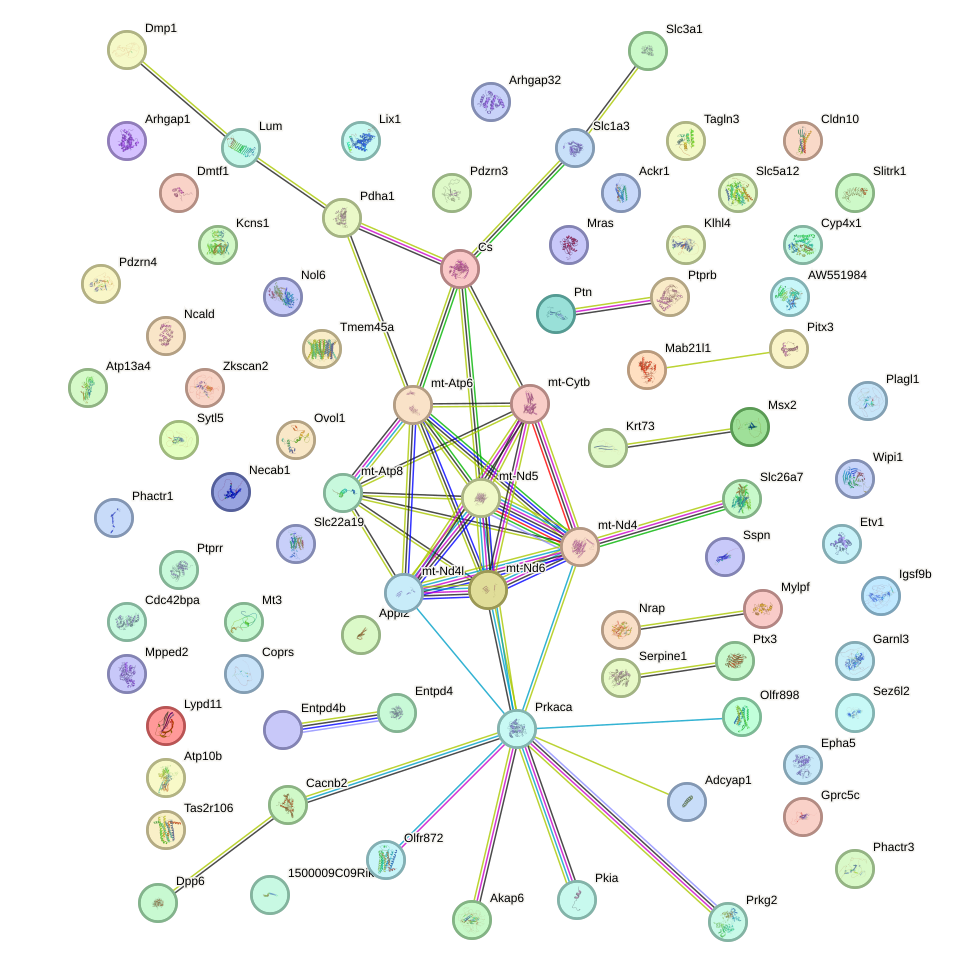
Network of associatons between targets according to the STRING database.
First level regulatory network of Gsx2_Hoxd3_Vax1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_94879235 | 7.62 |
ENSMUST00000034211.10
ENSMUST00000211930.2 ENSMUST00000211915.2 |
Mt3
|
metallothionein 3 |
| chr17_+_17622934 | 7.37 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr15_-_8739893 | 5.93 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr15_-_8740218 | 5.49 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr4_-_14621805 | 5.14 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr9_+_32027335 | 4.89 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr5_-_99091681 | 3.95 |
ENSMUST00000162619.8
ENSMUST00000162147.6 |
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr2_+_91480513 | 3.92 |
ENSMUST00000090614.11
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr12_+_52746158 | 3.66 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chrX_+_9751861 | 3.51 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr2_+_91480460 | 3.39 |
ENSMUST00000111331.9
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr16_-_45544960 | 3.34 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr9_+_38738911 | 3.31 |
ENSMUST00000051238.7
ENSMUST00000219798.2 |
Olfr923
|
olfactory receptor 923 |
| chr12_+_38831093 | 3.20 |
ENSMUST00000161513.9
|
Etv1
|
ets variant 1 |
| chr4_-_14621497 | 3.07 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr19_-_5610628 | 2.96 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr1_+_179936757 | 2.79 |
ENSMUST00000143176.8
ENSMUST00000135056.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr14_+_65504067 | 2.64 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr5_-_84565218 | 2.56 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr15_-_37459570 | 2.55 |
ENSMUST00000119730.8
ENSMUST00000120746.8 |
Ncald
|
neurocalcin delta |
| chr17_+_85335775 | 2.44 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr12_+_38830812 | 2.39 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr13_-_53627110 | 2.32 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr11_-_109502243 | 2.21 |
ENSMUST00000103060.10
ENSMUST00000047186.10 ENSMUST00000106689.2 |
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr11_+_114741948 | 2.21 |
ENSMUST00000133245.2
ENSMUST00000122967.3 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr2_+_14828903 | 2.14 |
ENSMUST00000193800.6
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr11_+_60956624 | 2.07 |
ENSMUST00000041944.9
ENSMUST00000108717.3 |
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr6_-_36787096 | 2.05 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chr11_+_43046476 | 2.00 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr19_-_7688628 | 1.92 |
ENSMUST00000025666.8
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr12_+_38830283 | 1.89 |
ENSMUST00000162563.8
ENSMUST00000161164.8 ENSMUST00000160996.8 |
Etv1
|
ets variant 1 |
| chrX_-_100129626 | 1.84 |
ENSMUST00000113710.8
|
Slc7a3
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 |
| chr4_-_14621669 | 1.84 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr19_-_40371016 | 1.81 |
ENSMUST00000225766.3
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr10_-_129965752 | 1.77 |
ENSMUST00000215217.2
ENSMUST00000214192.2 |
Olfr824
|
olfactory receptor 824 |
| chr13_+_43019718 | 1.72 |
ENSMUST00000131942.2
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr3_+_7494108 | 1.72 |
ENSMUST00000193330.2
|
Pkia
|
protein kinase inhibitor, alpha |
| chr2_+_110427643 | 1.68 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr2_-_91480096 | 1.67 |
ENSMUST00000099714.10
ENSMUST00000111333.2 |
Zfp408
|
zinc finger protein 408 |
| chr15_+_82140224 | 1.65 |
ENSMUST00000143238.2
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr8_-_13940234 | 1.63 |
ENSMUST00000033839.9
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr1_-_83016152 | 1.62 |
ENSMUST00000164473.2
ENSMUST00000045560.15 |
Slc19a3
|
solute carrier family 19, member 3 |
| chr2_-_88559941 | 1.60 |
ENSMUST00000099815.2
|
Olfr1197
|
olfactory receptor 1197 |
| chr5_+_104350475 | 1.58 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr10_+_115653152 | 1.57 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr2_+_85715984 | 1.52 |
ENSMUST00000213441.3
|
Olfr1023
|
olfactory receptor 1023 |
| chr9_-_39514931 | 1.51 |
ENSMUST00000119722.8
|
AW551984
|
expressed sequence AW551984 |
| chr18_+_59195534 | 1.47 |
ENSMUST00000058633.9
ENSMUST00000175897.8 ENSMUST00000118510.8 ENSMUST00000175830.2 |
Minar2
|
membrane integral NOTCH2 associated receptor 2 |
| chr3_+_55689921 | 1.41 |
ENSMUST00000075422.6
|
Mab21l1
|
mab-21-like 1 |
| chr3_+_66127330 | 1.41 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chr15_-_101710781 | 1.41 |
ENSMUST00000063292.8
|
Krt73
|
keratin 73 |
| chr10_+_97400990 | 1.35 |
ENSMUST00000038160.6
|
Lum
|
lumican |
| chr6_+_145879839 | 1.33 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr15_+_38869667 | 1.32 |
ENSMUST00000022906.8
|
Fzd6
|
frizzled class receptor 6 |
| chr18_+_37827413 | 1.32 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr4_-_15149755 | 1.31 |
ENSMUST00000108273.2
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chrM_+_10167 | 1.31 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chrX_-_142716200 | 1.29 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr9_+_27210500 | 1.29 |
ENSMUST00000214357.2
ENSMUST00000115247.8 ENSMUST00000133213.3 |
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr11_-_73217633 | 1.28 |
ENSMUST00000134079.2
|
Aspa
|
aspartoacylase |
| chr18_+_59195354 | 1.28 |
ENSMUST00000165666.9
|
Minar2
|
membrane integral NOTCH2 associated receptor 2 |
| chrM_+_7758 | 1.27 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr19_-_56378309 | 1.27 |
ENSMUST00000166203.2
ENSMUST00000167239.8 |
Nrap
|
nebulin-related anchoring protein |
| chr10_+_128173603 | 1.25 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase |
| chr4_-_114991478 | 1.25 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr6_-_131655849 | 1.21 |
ENSMUST00000076756.3
|
Tas2r106
|
taste receptor, type 2, member 106 |
| chr15_+_38869415 | 1.20 |
ENSMUST00000179165.9
|
Fzd6
|
frizzled class receptor 6 |
| chr2_+_106523532 | 1.19 |
ENSMUST00000111063.8
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr9_+_20148415 | 1.19 |
ENSMUST00000086474.6
|
Olfr872
|
olfactory receptor 872 |
| chr2_-_164013033 | 1.19 |
ENSMUST00000045196.4
|
Kcns1
|
K+ voltage-gated channel, subfamily S, 1 |
| chr1_-_173161069 | 1.18 |
ENSMUST00000038227.6
|
Ackr1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr4_-_114991174 | 1.18 |
ENSMUST00000051400.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr10_-_83484467 | 1.17 |
ENSMUST00000146876.9
ENSMUST00000176294.2 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr16_-_56706494 | 1.17 |
ENSMUST00000023435.6
|
Tmem45a
|
transmembrane protein 45a |
| chr15_+_98468885 | 1.15 |
ENSMUST00000023728.8
|
Tex49
|
testis expressed 49 |
| chr2_-_32977182 | 1.15 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr10_+_12936248 | 1.14 |
ENSMUST00000193426.6
|
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chrM_-_14061 | 1.14 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr6_-_101354858 | 1.11 |
ENSMUST00000075994.11
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chrM_+_7779 | 1.10 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chrM_+_14138 | 1.08 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr1_+_135768409 | 1.07 |
ENSMUST00000189826.7
|
Tnnt2
|
troponin T2, cardiac |
| chr9_+_38260381 | 1.06 |
ENSMUST00000076504.2
|
Olfr898
|
olfactory receptor 898 |
| chr7_+_126549692 | 1.05 |
ENSMUST00000106335.8
ENSMUST00000146017.3 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chrM_+_9870 | 1.05 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr13_-_21257469 | 1.05 |
ENSMUST00000058168.2
|
Olfr1370
|
olfactory receptor 1370 |
| chr5_+_27022355 | 1.04 |
ENSMUST00000071500.13
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr19_+_12647803 | 1.04 |
ENSMUST00000207341.3
ENSMUST00000208494.3 ENSMUST00000208657.3 |
Olfr1442
|
olfactory receptor 1442 |
| chr14_+_119092107 | 1.03 |
ENSMUST00000100314.4
|
Cldn10
|
claudin 10 |
| chr15_+_92495007 | 1.03 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr18_+_12874390 | 1.02 |
ENSMUST00000121018.8
ENSMUST00000119108.8 ENSMUST00000186263.2 ENSMUST00000191078.7 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr9_-_99302205 | 1.02 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chrX_-_158921370 | 1.02 |
ENSMUST00000033662.9
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chrX_+_113384297 | 1.01 |
ENSMUST00000133447.2
|
Klhl4
|
kelch-like 4 |
| chr16_-_29363671 | 1.01 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr14_-_109151590 | 1.00 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr2_+_177760959 | 0.97 |
ENSMUST00000108916.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr7_+_126808016 | 0.97 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr7_-_24423715 | 0.97 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr11_-_65160810 | 0.95 |
ENSMUST00000108695.9
|
Myocd
|
myocardin |
| chr17_+_93506435 | 0.94 |
ENSMUST00000234646.2
ENSMUST00000234081.2 |
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr5_-_137101108 | 0.94 |
ENSMUST00000077523.4
ENSMUST00000041388.11 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr8_+_36956345 | 0.92 |
ENSMUST00000171777.2
|
Trmt9b
|
tRNA methyltransferase 9B |
| chr11_-_65160767 | 0.92 |
ENSMUST00000102635.10
|
Myocd
|
myocardin |
| chr14_+_69585036 | 0.92 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr10_+_115979787 | 0.91 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr8_-_41494890 | 0.90 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr7_-_123099672 | 0.89 |
ENSMUST00000042470.14
ENSMUST00000128217.2 |
Zkscan2
|
zinc finger with KRAB and SCAN domains 2 |
| chr10_+_116111441 | 0.88 |
ENSMUST00000218553.2
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr1_+_78286946 | 0.88 |
ENSMUST00000036172.10
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr10_+_32959472 | 0.88 |
ENSMUST00000095762.5
ENSMUST00000218281.2 ENSMUST00000217779.2 ENSMUST00000219665.2 ENSMUST00000219931.2 |
Trdn
|
triadin |
| chr17_+_93506590 | 0.87 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr11_+_59197746 | 0.86 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr9_-_50571080 | 0.86 |
ENSMUST00000034567.4
|
Dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr7_-_55669702 | 0.86 |
ENSMUST00000052204.6
|
Nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 homolog (human) |
| chr6_-_136852792 | 0.85 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr5_+_150218847 | 0.83 |
ENSMUST00000239118.2
|
Fry
|
FRY microtubule binding protein |
| chr2_+_20742115 | 0.83 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr19_-_38807600 | 0.83 |
ENSMUST00000025963.8
|
Noc3l
|
NOC3 like DNA replication regulator |
| chr2_+_69727599 | 0.82 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr10_+_115854118 | 0.80 |
ENSMUST00000063470.11
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr9_+_121780054 | 0.77 |
ENSMUST00000043011.9
ENSMUST00000214536.3 ENSMUST00000215990.3 |
Gask1a
|
golgi associated kinase 1A |
| chr3_+_138019040 | 0.75 |
ENSMUST00000013455.13
ENSMUST00000106247.2 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr16_-_16418397 | 0.75 |
ENSMUST00000159542.8
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr9_+_37119472 | 0.75 |
ENSMUST00000034632.10
|
Tmem218
|
transmembrane protein 218 |
| chr3_+_159545309 | 0.75 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr11_-_49004584 | 0.74 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr13_-_4659120 | 0.73 |
ENSMUST00000091848.7
ENSMUST00000110691.10 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr11_-_73217298 | 0.73 |
ENSMUST00000155630.9
|
Aspa
|
aspartoacylase |
| chr13_+_33579744 | 0.73 |
ENSMUST00000050276.9
|
Serpinb9h
|
serine (or cysteine) peptidase inhibitor, clade B, member 9h |
| chr9_+_109881083 | 0.73 |
ENSMUST00000164930.8
ENSMUST00000199498.5 |
Map4
|
microtubule-associated protein 4 |
| chrX_+_151909893 | 0.73 |
ENSMUST00000163801.2
|
Foxr2
|
forkhead box R2 |
| chr7_+_107702486 | 0.72 |
ENSMUST00000104917.4
|
Olfr483
|
olfactory receptor 483 |
| chr11_+_35011953 | 0.72 |
ENSMUST00000069837.4
|
Slit3
|
slit guidance ligand 3 |
| chr3_+_63148887 | 0.70 |
ENSMUST00000194324.6
|
Mme
|
membrane metallo endopeptidase |
| chr8_-_55177510 | 0.70 |
ENSMUST00000175915.8
|
Wdr17
|
WD repeat domain 17 |
| chr8_+_46111703 | 0.70 |
ENSMUST00000134675.8
ENSMUST00000139869.8 ENSMUST00000126067.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr2_+_177760768 | 0.69 |
ENSMUST00000108917.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr2_+_19376447 | 0.69 |
ENSMUST00000023856.9
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr1_-_4563821 | 0.68 |
ENSMUST00000191939.2
|
Sox17
|
SRY (sex determining region Y)-box 17 |
| chr7_-_103718362 | 0.66 |
ENSMUST00000077417.3
|
Olfr644
|
olfactory receptor 644 |
| chr5_-_143279378 | 0.66 |
ENSMUST00000212715.2
|
Zfp853
|
zinc finger protein 853 |
| chr1_+_34160331 | 0.66 |
ENSMUST00000183006.5
|
Dst
|
dystonin |
| chr2_+_69727563 | 0.66 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_+_85606930 | 0.64 |
ENSMUST00000076250.2
|
Olfr1014
|
olfactory receptor 1014 |
| chr7_-_10292412 | 0.64 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68 |
| chr13_-_23041731 | 0.64 |
ENSMUST00000228645.2
|
Vmn1r211
|
vomeronasal 1 receptor 211 |
| chr4_+_108576846 | 0.63 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr8_-_35432783 | 0.63 |
ENSMUST00000033929.6
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr11_+_114742331 | 0.62 |
ENSMUST00000177952.8
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr7_-_13856967 | 0.62 |
ENSMUST00000098809.4
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr9_-_99450693 | 0.62 |
ENSMUST00000185524.7
ENSMUST00000186049.2 |
Armc8
|
armadillo repeat containing 8 |
| chr19_+_34078333 | 0.61 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chrX_+_92718695 | 0.61 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr6_-_123395075 | 0.61 |
ENSMUST00000172199.3
|
Vmn2r20
|
vomeronasal 2, receptor 20 |
| chr5_-_86616849 | 0.60 |
ENSMUST00000101073.3
|
Tmprss11a
|
transmembrane protease, serine 11a |
| chr16_-_89615225 | 0.60 |
ENSMUST00000164263.9
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr16_-_4698148 | 0.59 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr8_-_112737952 | 0.59 |
ENSMUST00000034426.14
|
Kars
|
lysyl-tRNA synthetase |
| chr16_-_92196954 | 0.59 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr16_-_19341016 | 0.58 |
ENSMUST00000214315.2
|
Olfr167
|
olfactory receptor 167 |
| chr18_+_37898633 | 0.57 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr6_+_92793440 | 0.57 |
ENSMUST00000057977.4
|
A730049H05Rik
|
RIKEN cDNA A730049H05 gene |
| chr14_-_52628228 | 0.57 |
ENSMUST00000078171.2
|
Olfr1511
|
olfactory receptor 1511 |
| chr18_+_31742565 | 0.57 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr3_-_131096792 | 0.56 |
ENSMUST00000200236.2
ENSMUST00000106337.7 |
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chrX_+_168468186 | 0.55 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr1_+_134333720 | 0.55 |
ENSMUST00000173908.8
|
Cyb5r1
|
cytochrome b5 reductase 1 |
| chr10_-_75946790 | 0.55 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr10_+_75983285 | 0.54 |
ENSMUST00000020450.4
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr10_-_128918779 | 0.53 |
ENSMUST00000213579.2
|
Olfr767
|
olfactory receptor 767 |
| chr5_-_137015683 | 0.53 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr6_-_93769426 | 0.52 |
ENSMUST00000204788.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr10_+_127919142 | 0.51 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr7_+_51528788 | 0.50 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr1_+_75412646 | 0.50 |
ENSMUST00000131545.8
ENSMUST00000141124.2 |
Gmppa
|
GDP-mannose pyrophosphorylase A |
| chr11_+_73489420 | 0.50 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chr8_-_112737898 | 0.49 |
ENSMUST00000093120.13
ENSMUST00000164470.3 ENSMUST00000211990.2 |
Kars
|
lysyl-tRNA synthetase |
| chr19_+_11674413 | 0.48 |
ENSMUST00000069760.13
ENSMUST00000119053.3 |
Oosp3
|
oocyte secreted protein 3 |
| chr13_+_23433408 | 0.48 |
ENSMUST00000091719.2
|
Vmn1r223
|
vomeronasal 1 receptor 223 |
| chr5_+_67125759 | 0.48 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chr14_+_25980463 | 0.48 |
ENSMUST00000173155.8
|
Duxbl1
|
double homeobox B-like 1 |
| chr6_-_56546088 | 0.47 |
ENSMUST00000203372.3
|
Pde1c
|
phosphodiesterase 1C |
| chr6_+_134617903 | 0.47 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chrX_-_98514278 | 0.47 |
ENSMUST00000113797.4
ENSMUST00000113790.8 ENSMUST00000036354.7 ENSMUST00000167246.2 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chr17_-_79076487 | 0.47 |
ENSMUST00000233850.2
|
Heatr5b
|
HEAT repeat containing 5B |
| chr2_-_152422220 | 0.47 |
ENSMUST00000053180.4
|
Defb19
|
defensin beta 19 |
| chr5_+_137015873 | 0.46 |
ENSMUST00000004968.11
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr7_+_43885573 | 0.46 |
ENSMUST00000223070.2
ENSMUST00000205530.2 |
Gm36864
|
predicted gene, 36864 |
| chr19_-_32173824 | 0.46 |
ENSMUST00000151822.2
|
Sgms1
|
sphingomyelin synthase 1 |
| chr3_-_120965327 | 0.45 |
ENSMUST00000170781.2
ENSMUST00000039761.12 ENSMUST00000106467.8 ENSMUST00000106466.10 ENSMUST00000164925.9 |
Rwdd3
|
RWD domain containing 3 |
| chr17_+_20639425 | 0.45 |
ENSMUST00000170076.2
|
Vmn1r224
|
vomeronasal 1 receptor 224 |
| chr2_+_91287846 | 0.45 |
ENSMUST00000028689.4
|
Lrp4
|
low density lipoprotein receptor-related protein 4 |
| chr3_-_144680801 | 0.45 |
ENSMUST00000029923.10
ENSMUST00000238960.2 |
Clca4a
|
chloride channel accessory 4A |
| chr2_-_29677634 | 0.44 |
ENSMUST00000177467.8
ENSMUST00000113807.10 |
Trub2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr1_-_4479445 | 0.43 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr10_+_119828821 | 0.42 |
ENSMUST00000105261.9
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr9_-_112016966 | 0.41 |
ENSMUST00000178410.2
ENSMUST00000172380.10 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr5_-_3697806 | 0.41 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chrX_-_52328963 | 0.40 |
ENSMUST00000074861.9
|
Plac1
|
placental specific protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 2.3 | 11.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.2 | 7.3 | GO:0032439 | endosome localization(GO:0032439) negative regulation of vacuolar transport(GO:1903336) |
| 1.2 | 4.8 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 1.1 | 10.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 1.0 | 4.0 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 1.0 | 3.0 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.6 | 2.5 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.6 | 1.9 | GO:2000722 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.6 | 3.7 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.5 | 2.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.5 | 1.4 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.4 | 7.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.4 | 0.7 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.4 | 1.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.4 | 1.8 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.4 | 1.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.3 | 1.6 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.3 | 1.6 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.3 | 0.9 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 1.8 | GO:0015819 | lysine transport(GO:0015819) |
| 0.3 | 2.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.3 | 1.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.3 | 1.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 1.9 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.2 | 1.6 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 1.9 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.2 | 0.8 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 0.6 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 0.6 | GO:1904268 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.2 | 0.7 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.2 | 7.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 0.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.7 | GO:0003142 | cardiogenic plate morphogenesis(GO:0003142) regulation of transcription from RNA polymerase II promoter involved in definitive endodermal cell fate specification(GO:0060807) |
| 0.2 | 1.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 2.6 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 | 0.5 | GO:1902071 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.9 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 1.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 2.2 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.1 | 0.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.3 | GO:0009107 | lipoate metabolic process(GO:0009106) lipoate biosynthetic process(GO:0009107) |
| 0.1 | 0.7 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.5 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 1.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.1 | 1.5 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 0.3 | GO:0042374 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 1.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 2.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.7 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 2.8 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.2 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.1 | 3.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.2 | GO:0018931 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.1 | 0.4 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.5 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.2 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) late endosomal microautophagy(GO:0061738) |
| 0.0 | 1.4 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.9 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 2.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.8 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.3 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 1.2 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 1.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 1.1 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.9 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 1.1 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.2 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.6 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 1.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.6 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 1.0 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 | 0.9 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.3 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 2.6 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 1.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.5 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 2.1 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.0 | 7.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.3 | 1.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 0.9 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 0.9 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.2 | 1.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 1.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 0.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 1.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 2.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 2.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 1.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.1 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 1.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 2.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.5 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.5 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 1.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 3.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 15.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 3.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 1.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 9.8 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 2.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 5.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 1.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.8 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 2.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 3.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 3.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.5 | GO:0030427 | site of polarized growth(GO:0030427) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.7 | 2.8 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.7 | 2.0 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.7 | 4.0 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.6 | 10.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.5 | 2.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 1.6 | GO:0015563 | thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.4 | 1.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.3 | 1.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.3 | 2.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 0.9 | GO:0016418 | pyruvate dehydrogenase activity(GO:0004738) S-acetyltransferase activity(GO:0016418) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.3 | 2.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 0.9 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 3.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 0.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 1.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 1.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.1 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 3.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 1.9 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 3.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.9 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.7 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.6 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 0.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 1.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 3.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.3 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 1.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 6.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 2.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 2.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.0 | 0.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.4 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.5 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 1.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 8.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 1.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.1 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 12.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 2.3 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 2.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 4.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 11.9 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 3.4 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 7.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 5.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 2.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.9 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 15.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 2.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 9.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 3.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.1 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 4.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |