Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
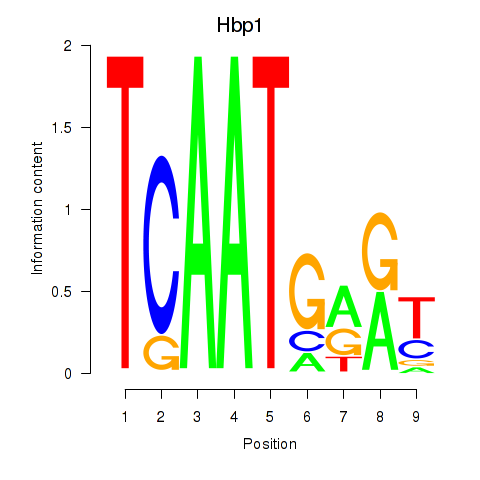
Results for Hbp1
Z-value: 1.90
Transcription factors associated with Hbp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hbp1
|
ENSMUSG00000002996.18 | Hbp1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hbp1 | mm39_v1_chr12_-_32000534_32000558 | -0.42 | 2.2e-04 | Click! |
Activity profile of Hbp1 motif
Sorted Z-values of Hbp1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hbp1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_115353326 | 24.19 |
ENSMUST00000030487.3
|
Cyp4a14
|
cytochrome P450, family 4, subfamily a, polypeptide 14 |
| chrX_+_10118544 | 21.67 |
ENSMUST00000049910.13
|
Otc
|
ornithine transcarbamylase |
| chr5_-_87572060 | 18.28 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr1_+_67162176 | 18.16 |
ENSMUST00000027144.8
|
Cps1
|
carbamoyl-phosphate synthetase 1 |
| chrX_+_10118600 | 18.05 |
ENSMUST00000115528.3
|
Otc
|
ornithine transcarbamylase |
| chr13_+_4486105 | 17.18 |
ENSMUST00000156277.2
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr19_+_40078132 | 17.12 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr5_+_90708962 | 16.37 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr5_-_87054796 | 15.89 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr19_-_39729431 | 15.46 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr19_+_39980868 | 15.38 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr19_-_44017637 | 14.28 |
ENSMUST00000026211.10
ENSMUST00000211830.2 |
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr13_-_63036096 | 12.91 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr3_+_146302832 | 12.26 |
ENSMUST00000029837.14
ENSMUST00000147409.2 ENSMUST00000121133.2 |
Uox
|
urate oxidase |
| chr5_-_145816774 | 12.10 |
ENSMUST00000035918.8
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr5_+_87148697 | 12.06 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr14_-_30665232 | 11.83 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr10_-_25412010 | 11.70 |
ENSMUST00000179685.3
|
Smlr1
|
small leucine-rich protein 1 |
| chr9_-_44714263 | 11.66 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr8_-_41668182 | 11.25 |
ENSMUST00000034003.5
|
Fgl1
|
fibrinogen-like protein 1 |
| chr19_-_7943365 | 10.63 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr5_-_147259245 | 10.63 |
ENSMUST00000100433.5
|
Urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5) decarboxylase |
| chr1_+_88022776 | 10.47 |
ENSMUST00000150634.8
ENSMUST00000058237.14 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr19_+_39275518 | 10.39 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr10_+_93324624 | 10.21 |
ENSMUST00000129421.8
|
Hal
|
histidine ammonia lyase |
| chr19_+_30210320 | 9.96 |
ENSMUST00000025797.7
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr1_+_93062962 | 9.81 |
ENSMUST00000027491.7
|
Agxt
|
alanine-glyoxylate aminotransferase |
| chr5_-_87240405 | 9.68 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr5_-_87074380 | 9.48 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr3_+_137983250 | 9.44 |
ENSMUST00000004232.10
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr13_+_93810911 | 9.41 |
ENSMUST00000048001.8
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr4_+_98919183 | 9.41 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr1_+_87998487 | 9.35 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr5_-_87288177 | 9.14 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr16_+_17149235 | 8.96 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr19_-_4489415 | 8.88 |
ENSMUST00000235680.2
ENSMUST00000117462.2 ENSMUST00000048197.10 |
Rhod
|
ras homolog family member D |
| chr4_-_61700450 | 8.73 |
ENSMUST00000107477.2
ENSMUST00000080606.9 |
Mup19
|
major urinary protein 19 |
| chr7_+_51530060 | 8.63 |
ENSMUST00000145049.2
|
Gas2
|
growth arrest specific 2 |
| chr7_-_19410749 | 8.53 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
| chr17_-_35351026 | 8.49 |
ENSMUST00000025249.7
|
Apom
|
apolipoprotein M |
| chr17_+_64907697 | 8.38 |
ENSMUST00000086723.10
|
Man2a1
|
mannosidase 2, alpha 1 |
| chr11_-_75329726 | 8.31 |
ENSMUST00000108437.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr2_-_34990689 | 8.30 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr19_-_39801188 | 7.88 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr19_+_38995463 | 7.87 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr8_+_105460627 | 7.86 |
ENSMUST00000034346.15
ENSMUST00000164182.3 |
Ces2a
|
carboxylesterase 2A |
| chr11_-_110142565 | 7.77 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr10_+_21253190 | 7.64 |
ENSMUST00000042699.14
|
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr11_-_43792013 | 7.64 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr7_+_43856724 | 7.32 |
ENSMUST00000077354.5
|
Klk1b4
|
kallikrein 1-related pepidase b4 |
| chr1_+_160806241 | 7.30 |
ENSMUST00000195760.2
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr1_-_136877277 | 7.29 |
ENSMUST00000168126.7
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr2_+_172994841 | 7.14 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr8_+_60958931 | 7.13 |
ENSMUST00000079472.4
|
Aadat
|
aminoadipate aminotransferase |
| chr12_-_81014849 | 7.07 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr3_-_10400710 | 6.93 |
ENSMUST00000078748.4
|
Slc10a5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5 |
| chr15_-_34495329 | 6.83 |
ENSMUST00000022946.6
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chr19_-_4548602 | 6.81 |
ENSMUST00000048482.8
|
2010003K11Rik
|
RIKEN cDNA 2010003K11 gene |
| chr7_-_19411866 | 6.78 |
ENSMUST00000142352.9
|
Apoc2
|
apolipoprotein C-II |
| chr1_+_171246593 | 6.63 |
ENSMUST00000171362.2
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr19_+_12610870 | 6.63 |
ENSMUST00000119960.2
|
Glyat
|
glycine-N-acyltransferase |
| chr2_+_58644922 | 6.47 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr9_+_74769166 | 6.43 |
ENSMUST00000056006.11
|
Onecut1
|
one cut domain, family member 1 |
| chr5_+_90666791 | 6.35 |
ENSMUST00000113179.9
ENSMUST00000128740.2 |
Afm
|
afamin |
| chr6_-_23132977 | 6.25 |
ENSMUST00000031707.14
|
Aass
|
aminoadipate-semialdehyde synthase |
| chr10_+_62897353 | 6.24 |
ENSMUST00000178684.3
ENSMUST00000020266.15 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr12_-_81014755 | 6.24 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr3_+_94280101 | 6.24 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr9_-_70842090 | 6.12 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr19_+_56276375 | 6.11 |
ENSMUST00000166049.8
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr11_-_75330302 | 6.10 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr19_-_8382424 | 6.09 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr1_+_88139678 | 6.00 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr6_-_71121324 | 5.81 |
ENSMUST00000074241.9
|
Thnsl2
|
threonine synthase-like 2 (bacterial) |
| chr19_+_56276343 | 5.76 |
ENSMUST00000095948.11
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr6_+_40619913 | 5.76 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr1_+_131725119 | 5.75 |
ENSMUST00000112393.9
ENSMUST00000048660.12 |
Pm20d1
|
peptidase M20 domain containing 1 |
| chr5_+_8943943 | 5.69 |
ENSMUST00000196067.2
|
Abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr8_+_70525546 | 5.61 |
ENSMUST00000110160.9
ENSMUST00000049197.6 |
Tm6sf2
|
transmembrane 6 superfamily member 2 |
| chr5_-_87402659 | 5.58 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr18_-_35760260 | 5.39 |
ENSMUST00000025212.8
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chr12_+_112073113 | 5.33 |
ENSMUST00000079400.6
|
Aspg
|
asparaginase |
| chr19_-_39451509 | 5.28 |
ENSMUST00000035488.3
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr1_+_88128323 | 5.24 |
ENSMUST00000049289.9
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr14_-_33996185 | 5.00 |
ENSMUST00000227006.2
|
Shld2
|
shieldin complex subunit 2 |
| chr7_+_86895851 | 4.90 |
ENSMUST00000032781.14
|
Nox4
|
NADPH oxidase 4 |
| chr1_+_21310821 | 4.87 |
ENSMUST00000121676.8
ENSMUST00000124990.3 |
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr1_+_21310803 | 4.78 |
ENSMUST00000027067.15
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr2_+_105054657 | 4.72 |
ENSMUST00000068813.3
|
Them7
|
thioesterase superfamily member 7 |
| chr15_-_96947963 | 4.64 |
ENSMUST00000230907.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr8_+_46081213 | 4.59 |
ENSMUST00000130850.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr1_+_87983099 | 4.46 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chrX_+_149330371 | 4.44 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr3_+_142300601 | 4.38 |
ENSMUST00000029936.5
|
Gbp2b
|
guanylate binding protein 2b |
| chr8_-_22193658 | 4.30 |
ENSMUST00000071886.7
|
Defa39
|
defensin, alpha, 39 |
| chr15_+_78915071 | 4.30 |
ENSMUST00000006544.9
ENSMUST00000171999.9 |
Gcat
Gcat
|
glycine C-acetyltransferase (2-amino-3-ketobutyrate-coenzyme A ligase) glycine C-acetyltransferase (2-amino-3-ketobutyrate-coenzyme A ligase) |
| chr9_-_48516447 | 4.29 |
ENSMUST00000034808.12
ENSMUST00000119426.2 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr1_+_58152295 | 4.25 |
ENSMUST00000040999.14
ENSMUST00000162011.3 |
Aox3
|
aldehyde oxidase 3 |
| chr10_+_87697155 | 4.20 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr9_-_70841881 | 4.05 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chr17_+_43581220 | 4.04 |
ENSMUST00000047399.6
|
Adgrf1
|
adhesion G protein-coupled receptor F1 |
| chr12_-_28673311 | 4.00 |
ENSMUST00000036136.9
|
Colec11
|
collectin sub-family member 11 |
| chr3_+_132335575 | 4.00 |
ENSMUST00000212804.2
ENSMUST00000212852.2 |
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr1_-_91340884 | 3.96 |
ENSMUST00000086851.2
|
Hes6
|
hairy and enhancer of split 6 |
| chr2_-_110136074 | 3.93 |
ENSMUST00000046233.9
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr7_+_86895996 | 3.89 |
ENSMUST00000068829.13
|
Nox4
|
NADPH oxidase 4 |
| chr6_+_137731526 | 3.78 |
ENSMUST00000203216.3
ENSMUST00000087675.9 ENSMUST00000203693.3 |
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr8_-_21586066 | 3.71 |
ENSMUST00000077452.4
|
Defa38
|
defensin, alpha, 38 |
| chr14_+_55798517 | 3.59 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr3_-_98417351 | 3.57 |
ENSMUST00000179429.6
|
Hsd3b4
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 4 |
| chr3_+_63203516 | 3.57 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chrX_+_138464065 | 3.55 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr10_-_24712034 | 3.54 |
ENSMUST00000218044.2
ENSMUST00000020169.9 |
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr9_-_96900876 | 3.52 |
ENSMUST00000055433.5
|
Spsb4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr7_+_114367971 | 3.50 |
ENSMUST00000117543.3
ENSMUST00000151464.2 |
Insc
|
INSC spindle orientation adaptor protein |
| chr3_-_102871440 | 3.47 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chrX_-_99638466 | 3.43 |
ENSMUST00000053373.2
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr11_-_43727071 | 3.39 |
ENSMUST00000167574.2
|
Adra1b
|
adrenergic receptor, alpha 1b |
| chr9_-_35010357 | 3.39 |
ENSMUST00000214526.2
ENSMUST00000217149.2 |
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr17_-_35081129 | 3.36 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr4_+_101276474 | 3.14 |
ENSMUST00000102780.8
ENSMUST00000106946.8 ENSMUST00000106945.8 |
Ak4
|
adenylate kinase 4 |
| chr8_-_118400418 | 3.14 |
ENSMUST00000173522.8
ENSMUST00000174450.2 |
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr13_+_4283729 | 3.13 |
ENSMUST00000081326.7
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr6_+_65929546 | 3.12 |
ENSMUST00000043382.9
|
4930544G11Rik
|
RIKEN cDNA 4930544G11 gene |
| chr3_-_107838895 | 3.09 |
ENSMUST00000133947.9
ENSMUST00000124215.2 ENSMUST00000106688.8 ENSMUST00000106687.9 |
Gstm7
|
glutathione S-transferase, mu 7 |
| chr18_-_43870622 | 3.09 |
ENSMUST00000025381.4
|
Spink1
|
serine peptidase inhibitor, Kazal type 1 |
| chr13_+_4099001 | 3.08 |
ENSMUST00000118717.10
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr5_-_108022900 | 3.06 |
ENSMUST00000138111.8
ENSMUST00000112642.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr4_+_109092610 | 3.05 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr8_+_21555054 | 3.04 |
ENSMUST00000078121.4
|
Defa35
|
defensin, alpha, 35 |
| chr6_+_78347636 | 2.99 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr12_+_59142439 | 2.99 |
ENSMUST00000219140.3
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr2_+_15531281 | 2.96 |
ENSMUST00000146205.3
|
Malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr3_+_20011405 | 2.91 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr1_-_183766195 | 2.90 |
ENSMUST00000050306.8
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene |
| chr15_+_4928624 | 2.89 |
ENSMUST00000045736.9
|
Mroh2b
|
maestro heat-like repeat family member 2B |
| chr9_-_98917700 | 2.83 |
ENSMUST00000076730.8
|
Gm1123
|
predicted gene 1123 |
| chr12_-_69771604 | 2.82 |
ENSMUST00000021370.10
|
L2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr14_+_52254308 | 2.80 |
ENSMUST00000100638.4
|
Tmem253
|
transmembrane protein 253 |
| chr3_+_107538638 | 2.80 |
ENSMUST00000106703.2
|
Gm10961
|
predicted gene 10961 |
| chr2_-_111100733 | 2.79 |
ENSMUST00000099619.6
|
Olfr1277
|
olfactory receptor 1277 |
| chr2_-_64806106 | 2.77 |
ENSMUST00000156765.2
|
Grb14
|
growth factor receptor bound protein 14 |
| chr4_+_116578117 | 2.74 |
ENSMUST00000045542.13
ENSMUST00000106459.8 |
Tesk2
|
testis-specific kinase 2 |
| chr3_-_88204286 | 2.68 |
ENSMUST00000107556.10
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr18_+_36661198 | 2.66 |
ENSMUST00000237174.2
ENSMUST00000236124.2 ENSMUST00000236779.2 ENSMUST00000235181.2 ENSMUST00000074298.13 ENSMUST00000115694.3 ENSMUST00000236126.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr1_+_155911136 | 2.63 |
ENSMUST00000111757.10
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr17_+_29487881 | 2.61 |
ENSMUST00000234845.2
ENSMUST00000235038.2 ENSMUST00000235050.2 ENSMUST00000120346.9 ENSMUST00000234377.2 ENSMUST00000235074.2 ENSMUST00000235040.2 ENSMUST00000234256.2 ENSMUST00000234459.2 |
BC004004
|
cDNA sequence BC004004 |
| chr1_+_87983189 | 2.61 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr2_-_29983618 | 2.60 |
ENSMUST00000081838.7
ENSMUST00000102865.11 |
Zdhhc12
|
zinc finger, DHHC domain containing 12 |
| chr5_-_30703180 | 2.59 |
ENSMUST00000065486.3
|
Cib4
|
calcium and integrin binding family member 4 |
| chr15_+_54975713 | 2.59 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr14_+_26722319 | 2.58 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr5_-_66330394 | 2.57 |
ENSMUST00000201544.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr17_-_29768531 | 2.56 |
ENSMUST00000168339.3
ENSMUST00000114683.10 ENSMUST00000234620.2 |
Tmem217
|
transmembrane protein 217 |
| chr8_-_21817031 | 2.55 |
ENSMUST00000098890.4
|
Defa29
|
defensin, alpha, 29 |
| chr7_-_98887770 | 2.51 |
ENSMUST00000064231.8
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr2_-_155434487 | 2.51 |
ENSMUST00000155347.2
ENSMUST00000130881.8 ENSMUST00000079691.13 |
Gss
|
glutathione synthetase |
| chr9_-_51240201 | 2.51 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr6_-_138056914 | 2.50 |
ENSMUST00000171804.4
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr2_-_52225146 | 2.48 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr9_-_65330231 | 2.47 |
ENSMUST00000065894.7
|
Slc51b
|
solute carrier family 51, beta subunit |
| chr17_-_34000804 | 2.46 |
ENSMUST00000002360.17
|
Angptl4
|
angiopoietin-like 4 |
| chr8_+_67943410 | 2.45 |
ENSMUST00000026677.4
|
Nat1
|
N-acetyl transferase 1 |
| chr8_+_21805562 | 2.44 |
ENSMUST00000167683.3
ENSMUST00000168340.2 |
Defa27
|
defensin, alpha, 27 |
| chrX_-_137985960 | 2.42 |
ENSMUST00000033626.15
ENSMUST00000060824.4 |
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr17_-_66901568 | 2.42 |
ENSMUST00000024914.4
|
Themis3
|
thymocyte selection associated family member 3 |
| chr2_-_89487877 | 2.40 |
ENSMUST00000099768.3
|
Olfr1250
|
olfactory receptor 1250 |
| chr6_+_54244116 | 2.40 |
ENSMUST00000114402.9
|
Chn2
|
chimerin 2 |
| chr17_+_47999916 | 2.38 |
ENSMUST00000156118.8
|
Frs3
|
fibroblast growth factor receptor substrate 3 |
| chr3_+_116306719 | 2.37 |
ENSMUST00000000349.11
ENSMUST00000197201.5 |
Dbt
|
dihydrolipoamide branched chain transacylase E2 |
| chr17_+_37269468 | 2.37 |
ENSMUST00000040177.7
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr5_-_135601887 | 2.32 |
ENSMUST00000004936.10
ENSMUST00000201401.2 |
Ccl24
|
chemokine (C-C motif) ligand 24 |
| chr11_-_75313412 | 2.31 |
ENSMUST00000138661.8
ENSMUST00000000769.14 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr11_-_48708159 | 2.29 |
ENSMUST00000047145.14
|
Trim41
|
tripartite motif-containing 41 |
| chr12_+_113038376 | 2.28 |
ENSMUST00000109729.3
|
Tex22
|
testis expressed gene 22 |
| chr12_+_78243846 | 2.26 |
ENSMUST00000188791.2
|
Gm6657
|
predicted gene 6657 |
| chr10_+_27950809 | 2.26 |
ENSMUST00000166468.2
ENSMUST00000218359.2 ENSMUST00000218276.2 |
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr1_+_132243849 | 2.25 |
ENSMUST00000072177.14
ENSMUST00000082125.6 |
Nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr2_+_71884943 | 2.23 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr9_+_27308497 | 2.23 |
ENSMUST00000214287.2
|
Spata19
|
spermatogenesis associated 19 |
| chr1_-_171359228 | 2.21 |
ENSMUST00000168184.2
|
Itln1
|
intelectin 1 (galactofuranose binding) |
| chr9_+_77661808 | 2.19 |
ENSMUST00000034905.9
|
Gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr3_-_88204145 | 2.18 |
ENSMUST00000010682.4
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr11_+_115331365 | 2.17 |
ENSMUST00000093914.5
|
Trim80
|
tripartite motif-containing 80 |
| chr2_+_153742294 | 2.17 |
ENSMUST00000088955.12
ENSMUST00000135501.3 |
Bpifb6
|
BPI fold containing family B, member 6 |
| chr9_+_54771064 | 2.15 |
ENSMUST00000034843.9
|
Ireb2
|
iron responsive element binding protein 2 |
| chr4_-_155445779 | 2.14 |
ENSMUST00000105624.2
|
Prkcz
|
protein kinase C, zeta |
| chr3_-_153650269 | 2.13 |
ENSMUST00000072697.13
|
Acadm
|
acyl-Coenzyme A dehydrogenase, medium chain |
| chr11_-_115310743 | 2.13 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr7_+_78922947 | 2.12 |
ENSMUST00000037315.13
|
Abhd2
|
abhydrolase domain containing 2 |
| chr17_-_45906428 | 2.11 |
ENSMUST00000171081.8
ENSMUST00000172301.8 ENSMUST00000167332.8 ENSMUST00000170488.8 ENSMUST00000167195.8 ENSMUST00000064889.13 ENSMUST00000051574.13 ENSMUST00000164217.8 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr7_-_16348862 | 2.11 |
ENSMUST00000171937.2
ENSMUST00000075845.11 |
Arhgap35
|
Rho GTPase activating protein 35 |
| chrX_+_6933733 | 2.07 |
ENSMUST00000057101.13
ENSMUST00000115750.8 |
Akap4
|
A kinase (PRKA) anchor protein 4 |
| chr6_-_83654789 | 2.07 |
ENSMUST00000037882.8
|
Cd207
|
CD207 antigen |
| chr14_+_69409251 | 2.07 |
ENSMUST00000062437.10
|
Nkx2-6
|
NK2 homeobox 6 |
| chr1_+_174218612 | 2.06 |
ENSMUST00000075329.3
|
Olfr248
|
olfactory receptor 248 |
| chr8_-_21946576 | 2.06 |
ENSMUST00000110752.4
|
Defa42
|
defensin, alpha, 42 |
| chr9_+_38399327 | 2.01 |
ENSMUST00000045493.2
|
Olfr906
|
olfactory receptor 906 |
| chr14_-_118370144 | 2.01 |
ENSMUST00000022727.10
ENSMUST00000228543.2 |
Tgds
|
TDP-glucose 4,6-dehydratase |
| chr2_+_102536701 | 2.01 |
ENSMUST00000123759.8
ENSMUST00000005220.11 ENSMUST00000111212.8 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.2 | 39.7 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 5.9 | 17.7 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 5.2 | 15.5 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 3.8 | 15.3 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 3.7 | 11.0 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 3.6 | 18.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 3.5 | 10.5 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 3.3 | 9.8 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) oxalic acid secretion(GO:0046724) |
| 3.2 | 12.9 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 3.2 | 9.6 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 2.8 | 8.5 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 2.4 | 80.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 2.4 | 21.7 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 2.4 | 9.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 2.1 | 6.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 2.1 | 6.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 2.0 | 10.2 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 2.0 | 10.2 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 2.0 | 11.9 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 1.9 | 5.7 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 1.8 | 14.4 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 1.8 | 7.1 | GO:0061402 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 1.8 | 5.3 | GO:0006530 | asparagine catabolic process(GO:0006530) |
| 1.5 | 6.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.3 | 3.8 | GO:0033189 | response to vitamin A(GO:0033189) |
| 1.2 | 11.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 1.2 | 5.8 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 1.1 | 3.4 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 1.1 | 16.7 | GO:0015747 | urate transport(GO:0015747) |
| 1.0 | 10.6 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.9 | 7.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.9 | 8.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.9 | 2.7 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.8 | 2.5 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.8 | 3.1 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.8 | 10.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.8 | 8.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.8 | 3.8 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.7 | 3.0 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.7 | 12.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.7 | 7.9 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.7 | 2.1 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.7 | 3.6 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.7 | 6.2 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.7 | 2.7 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.7 | 3.3 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.7 | 1.3 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.6 | 7.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.6 | 4.2 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.6 | 1.8 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.6 | 1.7 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.5 | 2.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.5 | 3.1 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.5 | 14.8 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.5 | 15.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 1.5 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.5 | 3.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.5 | 2.3 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.4 | 2.2 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.4 | 1.3 | GO:1904735 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 0.4 | 2.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.4 | 1.1 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.4 | 4.5 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.4 | 2.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.4 | 1.8 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.4 | 2.2 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.4 | 1.1 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.4 | 6.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 1.1 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.3 | 1.0 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.3 | 1.3 | GO:2000832 | testosterone secretion(GO:0035936) negative regulation of steroid hormone secretion(GO:2000832) regulation of testosterone secretion(GO:2000843) |
| 0.3 | 1.9 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.3 | 2.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.3 | 1.5 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.3 | 3.7 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.3 | 2.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.3 | 2.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.3 | 0.9 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.3 | 1.5 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.3 | 1.2 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.3 | 2.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 0.9 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.3 | 1.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 1.7 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.3 | 1.7 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 1.9 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.3 | 1.4 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.3 | 1.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.3 | 1.8 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.3 | 11.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.3 | 2.0 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.2 | 4.4 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.2 | 2.6 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.2 | 1.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 3.5 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.2 | 2.6 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 0.7 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.2 | 2.9 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 0.9 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.2 | 1.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.2 | 8.8 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.2 | 5.6 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.2 | 0.6 | GO:0002276 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.2 | 1.8 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 2.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 2.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 0.6 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.2 | 0.5 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.2 | 0.9 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.2 | 0.5 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.2 | 4.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 1.7 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 1.7 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 1.2 | GO:0046689 | response to glucagon(GO:0033762) response to mercury ion(GO:0046689) |
| 0.2 | 3.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 2.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 1.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 1.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 0.9 | GO:0097466 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.2 | 0.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 1.1 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 2.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 4.6 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 1.3 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 1.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.6 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.1 | 2.6 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 1.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.7 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.5 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 1.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.8 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 6.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.4 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 0.1 | 1.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 1.1 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 2.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.6 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 2.1 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 2.6 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 5.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.7 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 1.6 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 2.7 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 0.3 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.1 | 0.7 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 2.4 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.1 | 3.0 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 5.7 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 6.4 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.1 | 0.7 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.1 | 0.5 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 2.9 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.6 | GO:0070895 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.1 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 5.0 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 0.9 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.4 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 1.6 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 1.1 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.1 | 0.4 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.1 | 0.1 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.1 | 0.8 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.9 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 1.8 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.1 | 1.0 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 1.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 3.5 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 2.0 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 1.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.3 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.5 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.1 | 0.4 | GO:0048687 | astrocyte activation(GO:0048143) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 1.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.2 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.1 | 0.6 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.2 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 1.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 3.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 1.1 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 1.4 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 1.0 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 0.6 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.3 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 | 1.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.4 | GO:1903543 | regulation of exosomal secretion(GO:1903541) positive regulation of exosomal secretion(GO:1903543) |
| 0.1 | 1.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 2.3 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 0.7 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 1.2 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 1.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.9 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 1.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.3 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.4 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 1.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 14.1 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 2.2 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.3 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.7 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 1.0 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.5 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 1.1 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 1.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 1.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 23.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.5 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 2.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 2.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0044036 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.1 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 3.3 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.5 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.2 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.5 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 1.5 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.1 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.0 | 0.1 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.6 | GO:1901606 | alpha-amino acid catabolic process(GO:1901606) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 15.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.7 | 8.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.6 | 14.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.1 | 5.6 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.7 | 2.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.6 | 6.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.6 | 5.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.5 | 8.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.5 | 5.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.5 | 1.9 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.4 | 2.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.4 | 1.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.4 | 0.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 6.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.4 | 1.4 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.3 | 20.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.3 | 9.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 1.6 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.3 | 10.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 4.4 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 1.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 1.5 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.3 | 1.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.3 | 8.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 1.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.2 | 1.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 0.9 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 3.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.3 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.2 | 0.6 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.2 | 1.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 0.7 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 2.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 1.5 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 4.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.3 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 3.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.7 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.1 | 2.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 35.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 3.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.0 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 30.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 51.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 0.6 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.1 | 0.3 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.1 | 3.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 16.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.9 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 8.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 8.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.9 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 1.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.7 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 19.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 2.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 6.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 88.2 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 0.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 3.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 5.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 1.6 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 3.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.9 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 13.9 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.2 | 18.2 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 9.9 | 39.7 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 6.2 | 25.0 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 5.9 | 17.7 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 3.5 | 24.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 3.1 | 9.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 2.9 | 118.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 2.8 | 8.4 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 2.8 | 11.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 2.7 | 16.3 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 2.6 | 15.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 2.5 | 20.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 2.4 | 7.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 2.4 | 9.4 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 2.1 | 6.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 2.0 | 10.2 | GO:0035478 | chylomicron binding(GO:0035478) |
| 1.8 | 5.3 | GO:0004067 | asparaginase activity(GO:0004067) |
| 1.7 | 38.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.7 | 6.6 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 1.6 | 9.8 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 1.5 | 12.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 1.5 | 4.4 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 1.5 | 10.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.5 | 4.4 | GO:0019002 | GMP binding(GO:0019002) |
| 1.5 | 5.8 | GO:0070905 | serine binding(GO:0070905) |
| 1.2 | 10.0 | GO:0005534 | galactose binding(GO:0005534) |
| 1.1 | 3.4 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 1.1 | 3.4 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 1.1 | 6.5 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 1.0 | 3.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 1.0 | 6.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.0 | 3.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 1.0 | 7.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 1.0 | 16.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.0 | 2.9 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 1.0 | 5.8 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.9 | 5.7 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.9 | 12.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.9 | 9.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.8 | 8.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.8 | 2.3 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.8 | 6.8 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.7 | 11.9 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.7 | 2.2 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.7 | 2.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.7 | 3.5 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.7 | 1.3 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 0.5 | 6.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.5 | 2.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.5 | 2.5 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.5 | 3.4 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.5 | 1.4 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.4 | 1.3 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.4 | 6.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.4 | 2.5 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.4 | 2.7 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.4 | 1.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.4 | 5.8 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.3 | 1.0 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.3 | 3.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 1.3 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.3 | 5.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 0.9 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.3 | 3.7 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 1.2 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.3 | 3.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 1.4 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.3 | 45.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 1.8 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.3 | 1.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.3 | 0.8 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.3 | 1.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.2 | 0.7 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.2 | 1.5 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.2 | 1.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.2 | 1.9 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.2 | 1.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 1.1 | GO:0004904 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.2 | 12.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 2.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 3.8 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 1.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 1.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 9.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 0.6 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.2 | 1.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 4.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.6 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.2 | 1.7 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.2 | 1.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 2.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 0.8 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.2 | 1.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.2 | 0.6 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.6 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.4 | GO:0001962 | alpha-1,3-galactosyltransferase activity(GO:0001962) |
| 0.1 | 1.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.9 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.8 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.7 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.8 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.4 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.1 | 2.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.3 | GO:0009918 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.1 | 2.5 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 3.4 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 12.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.9 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 4.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.6 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.3 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 2.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 3.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 3.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.9 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 2.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 6.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.6 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 1.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.7 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 3.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 1.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 2.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 1.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.9 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 2.8 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 4.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 4.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 39.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 5.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 1.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 1.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 2.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.8 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.1 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 3.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 2.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 40.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.9 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 3.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 10.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 6.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 9.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 2.4 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 12.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 0.9 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 3.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 0.9 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 6.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.2 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 4.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 2.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.4 | 15.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 1.2 | 13.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.0 | 8.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.9 | 25.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.7 | 2.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.6 | 7.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 7.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 3.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.4 | 94.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.3 | 6.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 17.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 23.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.3 | 6.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 8.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 13.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 8.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 9.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 3.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 3.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 2.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 5.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 2.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 7.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 8.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 3.4 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.1 | 2.4 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.1 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 6.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 2.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 6.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 9.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |