Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Hes5_Hes7
Z-value: 0.69
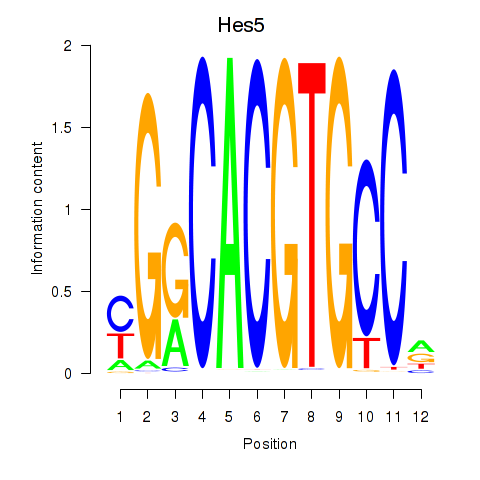
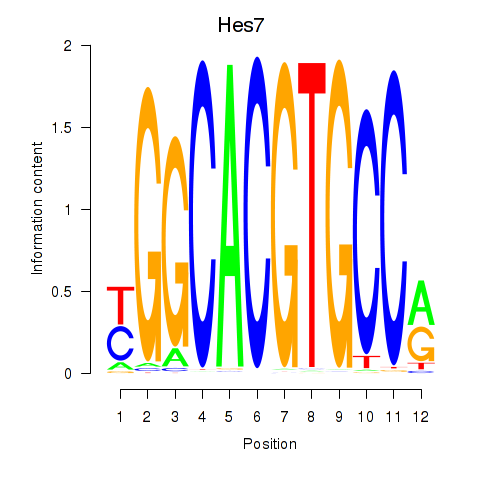
Transcription factors associated with Hes5_Hes7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hes5
|
ENSMUSG00000048001.8 | Hes5 |
|
Hes7
|
ENSMUSG00000023781.3 | Hes7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hes5 | mm39_v1_chr4_+_155045372_155045387 | 0.20 | 9.8e-02 | Click! |
| Hes7 | mm39_v1_chr11_+_69011230_69011230 | 0.19 | 1.2e-01 | Click! |
Activity profile of Hes5_Hes7 motif
Sorted Z-values of Hes5_Hes7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hes5_Hes7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_25438784 | 9.06 |
ENSMUST00000119720.8
ENSMUST00000121438.9 |
Adam32
|
a disintegrin and metallopeptidase domain 32 |
| chr3_-_95148909 | 8.54 |
ENSMUST00000090815.6
ENSMUST00000107197.2 |
Gm128
|
predicted gene 128 |
| chr9_-_121470564 | 7.36 |
ENSMUST00000213757.2
ENSMUST00000125075.2 ENSMUST00000077706.10 ENSMUST00000214592.2 |
Lyzl4
|
lysozyme-like 4 |
| chrX_+_8137372 | 6.16 |
ENSMUST00000127103.8
ENSMUST00000115591.8 |
Slc38a5
|
solute carrier family 38, member 5 |
| chrX_+_8137620 | 6.14 |
ENSMUST00000033512.11
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr19_-_5444413 | 5.73 |
ENSMUST00000044527.5
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr9_-_35469818 | 5.68 |
ENSMUST00000034612.7
|
Ddx25
|
DEAD box helicase 25 |
| chrX_+_8137881 | 5.64 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr4_+_134195631 | 4.67 |
ENSMUST00000030636.11
ENSMUST00000127279.8 ENSMUST00000105867.8 |
Stmn1
|
stathmin 1 |
| chr7_+_15795735 | 4.23 |
ENSMUST00000209369.2
|
Zfp541
|
zinc finger protein 541 |
| chr10_+_97528915 | 4.13 |
ENSMUST00000060703.6
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr2_+_155907658 | 3.84 |
ENSMUST00000137966.2
|
Spag4
|
sperm associated antigen 4 |
| chr17_-_26417982 | 3.78 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr7_+_43910845 | 3.54 |
ENSMUST00000124863.4
|
Gm15517
|
predicted gene 15517 |
| chr3_+_95466982 | 3.49 |
ENSMUST00000090797.11
ENSMUST00000171191.6 ENSMUST00000029754.13 ENSMUST00000107154.4 |
Hormad1
|
HORMA domain containing 1 |
| chr11_-_11976237 | 3.10 |
ENSMUST00000150972.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chr1_-_186875270 | 3.02 |
ENSMUST00000184543.2
|
Spata17
|
spermatogenesis associated 17 |
| chr9_-_107109108 | 2.78 |
ENSMUST00000044532.11
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr11_-_59678462 | 2.75 |
ENSMUST00000125307.2
|
Pld6
|
phospholipase D family, member 6 |
| chr8_+_120173458 | 2.72 |
ENSMUST00000098363.10
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr4_-_117109074 | 2.68 |
ENSMUST00000165128.9
|
Armh1
|
armadillo-like helical domain containing 1 |
| chr15_-_95426108 | 2.57 |
ENSMUST00000075275.3
|
Nell2
|
NEL-like 2 |
| chr6_+_72074545 | 2.40 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr4_+_154954042 | 2.39 |
ENSMUST00000079269.14
ENSMUST00000163732.8 ENSMUST00000080559.13 |
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr10_-_80223475 | 2.33 |
ENSMUST00000105350.3
|
Mex3d
|
mex3 RNA binding family member D |
| chr7_+_119927885 | 2.24 |
ENSMUST00000207220.2
ENSMUST00000121265.7 ENSMUST00000076272.5 |
Abca15
|
ATP-binding cassette, sub-family A (ABC1), member 15 |
| chr5_+_92534738 | 2.23 |
ENSMUST00000128246.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr12_-_72964534 | 2.13 |
ENSMUST00000110489.9
|
4930447C04Rik
|
RIKEN cDNA 4930447C04 gene |
| chr18_+_37276556 | 2.09 |
ENSMUST00000047479.3
|
Pcdhac2
|
protocadherin alpha subfamily C, 2 |
| chr6_+_35154545 | 1.89 |
ENSMUST00000170234.2
|
Nup205
|
nucleoporin 205 |
| chr2_-_130266162 | 1.74 |
ENSMUST00000089581.11
|
Pced1a
|
PC-esterase domain containing 1A |
| chr8_+_61085890 | 1.62 |
ENSMUST00000160719.8
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr8_+_61085853 | 1.60 |
ENSMUST00000161421.2
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr8_+_122996308 | 1.52 |
ENSMUST00000055537.3
|
Zfp469
|
zinc finger protein 469 |
| chrX_+_126301916 | 1.52 |
ENSMUST00000051530.4
|
4921511C20Rik
|
RIKEN cDNA 4921511C20 gene |
| chr17_-_3746536 | 1.51 |
ENSMUST00000115800.2
|
Nox3
|
NADPH oxidase 3 |
| chr6_+_72074718 | 1.44 |
ENSMUST00000187007.3
|
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr13_-_10410857 | 1.38 |
ENSMUST00000187510.7
|
Chrm3
|
cholinergic receptor, muscarinic 3, cardiac |
| chr5_-_46014809 | 1.36 |
ENSMUST00000190036.7
ENSMUST00000189859.7 ENSMUST00000186633.3 ENSMUST00000016026.14 ENSMUST00000045586.13 ENSMUST00000238522.2 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr10_-_67748461 | 1.30 |
ENSMUST00000064656.8
|
Zfp365
|
zinc finger protein 365 |
| chr9_+_45029080 | 1.28 |
ENSMUST00000170998.9
ENSMUST00000093855.4 |
Scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr5_+_28370687 | 1.28 |
ENSMUST00000036177.9
|
En2
|
engrailed 2 |
| chr1_+_9978863 | 1.27 |
ENSMUST00000052843.12
ENSMUST00000171802.8 ENSMUST00000125294.9 ENSMUST00000140948.9 |
Mcmdc2
|
minichromosome maintenance domain containing 2 |
| chr9_-_21874802 | 1.22 |
ENSMUST00000006397.7
|
Epor
|
erythropoietin receptor |
| chr1_-_161704224 | 1.21 |
ENSMUST00000048377.11
|
Suco
|
SUN domain containing ossification factor |
| chr9_-_78350486 | 1.16 |
ENSMUST00000070742.14
ENSMUST00000034898.14 |
Cgas
|
cyclic GMP-AMP synthase |
| chr13_+_49806542 | 1.09 |
ENSMUST00000222197.2
ENSMUST00000221083.2 ENSMUST00000223467.2 |
Nol8
|
nucleolar protein 8 |
| chr12_+_16703383 | 1.07 |
ENSMUST00000221596.2
ENSMUST00000111064.3 ENSMUST00000220892.2 |
Ntsr2
|
neurotensin receptor 2 |
| chr15_+_81954197 | 1.05 |
ENSMUST00000229119.2
ENSMUST00000089178.11 |
Mei1
|
meiotic double-stranded break formation protein 1 |
| chr6_+_91134358 | 1.02 |
ENSMUST00000155007.2
|
Hdac11
|
histone deacetylase 11 |
| chr4_-_149211145 | 1.00 |
ENSMUST00000030815.3
|
Cort
|
cortistatin |
| chr11_+_4587733 | 0.95 |
ENSMUST00000070257.14
ENSMUST00000109930.3 |
Ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr11_-_86698484 | 0.95 |
ENSMUST00000018569.14
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr8_-_88531018 | 0.94 |
ENSMUST00000165770.9
|
Zfp423
|
zinc finger protein 423 |
| chr4_+_155493668 | 0.89 |
ENSMUST00000123952.9
ENSMUST00000238423.2 ENSMUST00000238620.2 ENSMUST00000151083.8 |
Cfap74
|
cilia and flagella associated protein 74 |
| chr17_+_48623157 | 0.89 |
ENSMUST00000049614.13
|
B430306N03Rik
|
RIKEN cDNA B430306N03 gene |
| chr13_+_52000704 | 0.87 |
ENSMUST00000021903.3
|
Gadd45g
|
growth arrest and DNA-damage-inducible 45 gamma |
| chr17_-_24428351 | 0.85 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr19_+_47167259 | 0.84 |
ENSMUST00000111808.11
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr4_+_117109148 | 0.83 |
ENSMUST00000062824.12
|
Tmem53
|
transmembrane protein 53 |
| chr11_+_103540391 | 0.80 |
ENSMUST00000057870.4
|
Rprml
|
reprimo-like |
| chr4_+_32657105 | 0.80 |
ENSMUST00000071642.11
ENSMUST00000178134.2 |
Mdn1
|
midasin AAA ATPase 1 |
| chr4_+_117109204 | 0.80 |
ENSMUST00000125943.8
ENSMUST00000106434.8 |
Tmem53
|
transmembrane protein 53 |
| chr2_+_73102269 | 0.78 |
ENSMUST00000090813.6
|
Sp9
|
trans-acting transcription factor 9 |
| chr2_+_144441817 | 0.75 |
ENSMUST00000028917.7
|
Dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr1_-_75195127 | 0.73 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr18_+_37453427 | 0.73 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr12_+_16703709 | 0.72 |
ENSMUST00000221049.2
|
Ntsr2
|
neurotensin receptor 2 |
| chr13_+_49806772 | 0.70 |
ENSMUST00000223264.2
ENSMUST00000221142.2 ENSMUST00000222333.2 ENSMUST00000021824.8 |
Nol8
|
nucleolar protein 8 |
| chr15_-_78739717 | 0.69 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr2_-_102231208 | 0.62 |
ENSMUST00000102573.8
|
Trim44
|
tripartite motif-containing 44 |
| chr9_+_75221415 | 0.62 |
ENSMUST00000215875.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr7_-_65020655 | 0.62 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr17_+_36290743 | 0.62 |
ENSMUST00000087200.4
|
Gnl1
|
guanine nucleotide binding protein-like 1 |
| chr11_+_117545037 | 0.61 |
ENSMUST00000026658.13
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr7_-_65020955 | 0.61 |
ENSMUST00000102592.10
|
Tjp1
|
tight junction protein 1 |
| chr11_+_101207021 | 0.54 |
ENSMUST00000142640.8
ENSMUST00000019470.14 |
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr16_-_45544960 | 0.52 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr2_+_91357100 | 0.44 |
ENSMUST00000111338.10
|
Ckap5
|
cytoskeleton associated protein 5 |
| chr4_-_132484559 | 0.44 |
ENSMUST00000030709.9
|
Smpdl3b
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr17_+_36172210 | 0.43 |
ENSMUST00000074259.15
ENSMUST00000174873.2 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr5_-_120610828 | 0.41 |
ENSMUST00000052258.14
ENSMUST00000031594.13 |
Sdsl
|
serine dehydratase-like |
| chr1_-_188740023 | 0.41 |
ENSMUST00000085678.8
|
Kctd3
|
potassium channel tetramerisation domain containing 3 |
| chr4_+_123798690 | 0.40 |
ENSMUST00000106202.4
|
Mycbp
|
MYC binding protein |
| chr7_-_130964469 | 0.39 |
ENSMUST00000059438.11
|
2310057M21Rik
|
RIKEN cDNA 2310057M21 gene |
| chr2_-_65397850 | 0.37 |
ENSMUST00000238483.2
ENSMUST00000100069.9 |
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr1_-_192718064 | 0.34 |
ENSMUST00000215093.2
ENSMUST00000195354.6 |
Syt14
|
synaptotagmin XIV |
| chr10_+_79518002 | 0.32 |
ENSMUST00000020550.13
|
Cdc34
|
cell division cycle 34 |
| chr17_-_36290129 | 0.32 |
ENSMUST00000165613.9
ENSMUST00000173872.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr11_+_3913970 | 0.30 |
ENSMUST00000109985.8
ENSMUST00000020705.5 |
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr15_+_32244947 | 0.28 |
ENSMUST00000067458.7
|
Sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr12_-_56583582 | 0.28 |
ENSMUST00000001536.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr3_-_95789338 | 0.27 |
ENSMUST00000161994.2
|
Ciart
|
circadian associated repressor of transcription |
| chr15_+_38662158 | 0.27 |
ENSMUST00000022904.8
ENSMUST00000228820.2 |
Atp6v1c1
|
ATPase, H+ transporting, lysosomal V1 subunit C1 |
| chr2_-_164032521 | 0.27 |
ENSMUST00000051272.8
|
Wfdc12
|
WAP four-disulfide core domain 12 |
| chr2_-_65397809 | 0.25 |
ENSMUST00000066432.12
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr3_-_95789505 | 0.22 |
ENSMUST00000159863.2
ENSMUST00000159739.8 ENSMUST00000036418.10 ENSMUST00000161866.8 |
Ciart
|
circadian associated repressor of transcription |
| chr10_+_79518138 | 0.21 |
ENSMUST00000166603.2
ENSMUST00000219791.2 ENSMUST00000219930.2 ENSMUST00000218964.2 |
Cdc34
|
cell division cycle 34 |
| chr5_-_135773047 | 0.16 |
ENSMUST00000153399.2
|
Tmem120a
|
transmembrane protein 120A |
| chr8_+_125448867 | 0.16 |
ENSMUST00000034463.4
|
Arv1
|
ARV1 homolog, fatty acid homeostasis modulator |
| chr12_+_72807985 | 0.16 |
ENSMUST00000021514.10
|
Ppm1a
|
protein phosphatase 1A, magnesium dependent, alpha isoform |
| chr2_-_152306532 | 0.15 |
ENSMUST00000099206.3
|
Defb23
|
defensin beta 23 |
| chr11_+_59197746 | 0.14 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr5_-_99876898 | 0.13 |
ENSMUST00000146396.8
ENSMUST00000161148.2 ENSMUST00000161516.2 |
Rasgef1b
|
RasGEF domain family, member 1B |
| chr7_-_98831916 | 0.10 |
ENSMUST00000033001.6
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr9_+_55056648 | 0.10 |
ENSMUST00000121677.8
|
Ube2q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr17_-_15181491 | 0.09 |
ENSMUST00000024657.12
|
Phf10
|
PHD finger protein 10 |
| chr13_+_22477363 | 0.09 |
ENSMUST00000091735.2
|
Vmn1r196
|
vomeronasal 1 receptor 196 |
| chr6_+_116490474 | 0.08 |
ENSMUST00000218028.2
ENSMUST00000220134.2 |
Olfr212
|
olfactory receptor 212 |
| chr14_-_69945022 | 0.07 |
ENSMUST00000118374.8
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr1_+_92500847 | 0.06 |
ENSMUST00000074859.3
|
Olfr1413
|
olfactory receptor 1413 |
| chr1_-_192717958 | 0.05 |
ENSMUST00000016344.9
|
Syt14
|
synaptotagmin XIV |
| chr14_-_69944942 | 0.05 |
ENSMUST00000121142.4
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr2_-_155534295 | 0.03 |
ENSMUST00000041059.12
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr11_-_50216426 | 0.03 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr4_+_123798625 | 0.02 |
ENSMUST00000030400.14
|
Mycbp
|
MYC binding protein |
| chr12_+_77285770 | 0.00 |
ENSMUST00000062804.8
|
Fut8
|
fucosyltransferase 8 |
| chr8_+_13155621 | 0.00 |
ENSMUST00000016680.14
ENSMUST00000121426.2 |
Cul4a
|
cullin 4A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 1.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 1.0 | 17.9 | GO:0015816 | glycine transport(GO:0015816) |
| 0.9 | 2.8 | GO:0008358 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.9 | 2.7 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.9 | 3.5 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.7 | 2.1 | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) |
| 0.7 | 4.7 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.5 | 1.5 | GO:0009629 | response to gravity(GO:0009629) |
| 0.4 | 7.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.4 | 1.9 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.3 | 1.3 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.3 | 1.4 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 | 3.8 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 1.8 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 1.0 | GO:0000237 | leptotene(GO:0000237) |
| 0.1 | 2.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 1.9 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 5.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 1.2 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 1.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 1.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 2.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.8 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 3.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 2.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 2.0 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.0 | 0.5 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.3 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 1.2 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.6 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.0 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.2 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.9 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.8 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.8 | GO:0007520 | myoblast fusion(GO:0007520) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 5.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 2.1 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 1.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 7.4 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 3.8 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 4.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.3 | 17.9 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.7 | 2.7 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.6 | 1.8 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.4 | 7.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 3.8 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 1.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 3.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 0.8 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.2 | 1.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 2.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.4 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.1 | 1.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 1.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 6.3 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 8.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 2.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.7 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 2.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 2.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.6 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.6 | GO:0031402 | voltage-gated sodium channel activity(GO:0005248) sodium ion binding(GO:0031402) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 2.5 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 2.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 7.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 17.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.9 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 2.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |