Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Hey2
Z-value: 0.77
Transcription factors associated with Hey2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hey2
|
ENSMUSG00000019789.10 | Hey2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hey2 | mm39_v1_chr10_-_30718760_30718797 | 0.13 | 2.9e-01 | Click! |
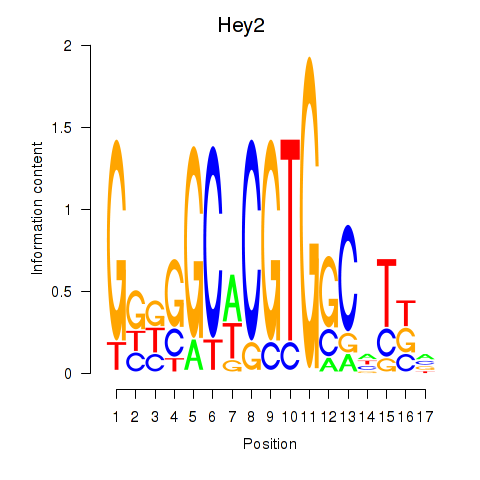
Activity profile of Hey2 motif
Sorted Z-values of Hey2 motif
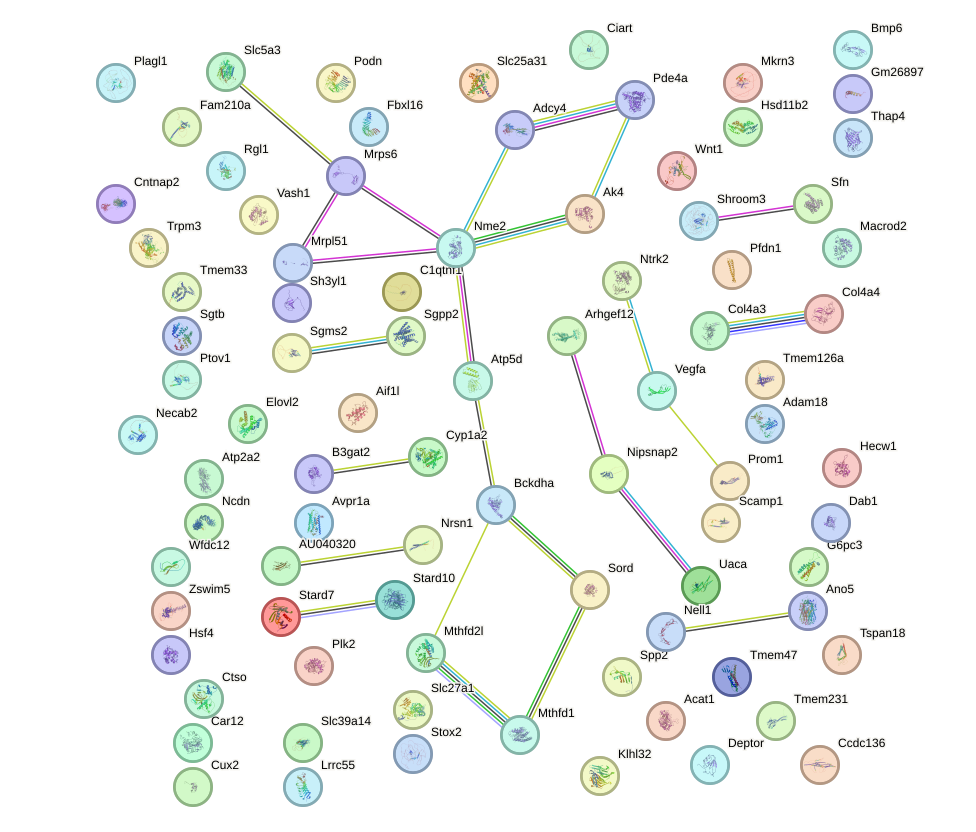
Network of associatons between targets according to the STRING database.
First level regulatory network of Hey2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_82564501 | 5.68 |
ENSMUST00000087050.7
|
Col4a4
|
collagen, type IV, alpha 4 |
| chr1_+_88334678 | 4.62 |
ENSMUST00000027518.12
|
Spp2
|
secreted phosphoprotein 2 |
| chr13_-_41373638 | 4.50 |
ENSMUST00000117096.2
|
Elovl2
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
| chr1_+_82564627 | 3.94 |
ENSMUST00000113457.9
|
Col4a3
|
collagen, type IV, alpha 3 |
| chr4_+_101276893 | 3.85 |
ENSMUST00000131397.2
ENSMUST00000133055.8 |
Ak4
|
adenylate kinase 4 |
| chr2_+_140237229 | 3.37 |
ENSMUST00000110067.8
ENSMUST00000110063.8 ENSMUST00000110064.8 ENSMUST00000110062.8 ENSMUST00000043836.8 ENSMUST00000078027.12 |
Macrod2
|
mono-ADP ribosylhydrolase 2 |
| chr7_-_25358406 | 3.25 |
ENSMUST00000071329.8
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr4_+_116734573 | 3.15 |
ENSMUST00000044823.4
|
Zswim5
|
zinc finger SWIM-type containing 5 |
| chr3_-_95789338 | 3.14 |
ENSMUST00000161994.2
|
Ciart
|
circadian associated repressor of transcription |
| chr7_-_30810422 | 2.81 |
ENSMUST00000039435.15
|
Hpn
|
hepsin |
| chr9_+_66620959 | 2.54 |
ENSMUST00000071889.13
|
Car12
|
carbonic anhydrase 12 |
| chr9_+_66621001 | 2.48 |
ENSMUST00000085420.12
|
Car12
|
carbonic anhydrase 12 |
| chr14_-_70588803 | 2.45 |
ENSMUST00000143153.2
ENSMUST00000127000.2 ENSMUST00000068044.14 ENSMUST00000022688.10 |
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr9_+_60701749 | 2.41 |
ENSMUST00000214354.2
ENSMUST00000050183.7 ENSMUST00000217656.2 |
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr2_+_31840151 | 2.36 |
ENSMUST00000001920.13
ENSMUST00000151276.3 |
Aif1l
|
allograft inflammatory factor 1-like |
| chr3_-_95789505 | 2.34 |
ENSMUST00000159863.2
ENSMUST00000159739.8 ENSMUST00000036418.10 ENSMUST00000161866.8 |
Ciart
|
circadian associated repressor of transcription |
| chr1_+_23801084 | 2.28 |
ENSMUST00000140583.2
|
B3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr1_-_175319842 | 2.24 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr12_+_86725459 | 2.09 |
ENSMUST00000021681.4
|
Vash1
|
vasohibin 1 |
| chr7_-_90106375 | 2.08 |
ENSMUST00000032844.7
|
Tmem126a
|
transmembrane protein 126A |
| chr11_-_93846453 | 2.07 |
ENSMUST00000072566.5
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr13_+_38529062 | 2.05 |
ENSMUST00000171970.3
|
Bmp6
|
bone morphogenetic protein 6 |
| chr5_-_122639840 | 2.05 |
ENSMUST00000177974.8
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chrX_+_80114242 | 2.03 |
ENSMUST00000171953.8
ENSMUST00000026760.3 |
Tmem47
|
transmembrane protein 47 |
| chr17_-_46342739 | 2.02 |
ENSMUST00000024747.14
|
Vegfa
|
vascular endothelial growth factor A |
| chr5_+_67418137 | 2.00 |
ENSMUST00000161369.3
|
Tmem33
|
transmembrane protein 33 |
| chr19_-_55087849 | 2.00 |
ENSMUST00000061856.6
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr2_+_122065230 | 1.92 |
ENSMUST00000110551.4
|
Sord
|
sorbitol dehydrogenase |
| chr7_+_100970435 | 1.91 |
ENSMUST00000210192.2
ENSMUST00000172630.8 |
Stard10
|
START domain containing 10 |
| chr7_+_100971034 | 1.86 |
ENSMUST00000173270.8
|
Stard10
|
START domain containing 10 |
| chr8_-_112660363 | 1.85 |
ENSMUST00000034429.9
|
Tmem231
|
transmembrane protein 231 |
| chr2_+_31840340 | 1.82 |
ENSMUST00000148056.4
|
Aif1l
|
allograft inflammatory factor 1-like |
| chr1_-_93659622 | 1.80 |
ENSMUST00000189728.7
|
Thap4
|
THAP domain containing 4 |
| chr18_-_68433398 | 1.80 |
ENSMUST00000042852.7
|
Fam210a
|
family with sequence similarity 210, member A |
| chr5_-_122640255 | 1.74 |
ENSMUST00000031423.10
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr17_+_26028059 | 1.73 |
ENSMUST00000045692.9
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr5_+_129802127 | 1.68 |
ENSMUST00000086046.10
ENSMUST00000186265.6 |
Nipsnap2
|
nipsnap homolog 2 |
| chr6_-_88852017 | 1.57 |
ENSMUST00000145944.3
|
Podxl2
|
podocalyxin-like 2 |
| chr6_-_88851579 | 1.56 |
ENSMUST00000061262.11
ENSMUST00000140455.8 ENSMUST00000145780.2 |
Podxl2
|
podocalyxin-like 2 |
| chr15_+_54975713 | 1.53 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr13_+_110531571 | 1.48 |
ENSMUST00000022212.9
|
Plk2
|
polo like kinase 2 |
| chr14_-_56021484 | 1.46 |
ENSMUST00000170223.9
ENSMUST00000002398.9 ENSMUST00000227031.2 |
Adcy4
|
adenylate cyclase 4 |
| chr15_+_54975814 | 1.45 |
ENSMUST00000100660.11
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr1_-_152642073 | 1.45 |
ENSMUST00000111857.3
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr1_+_23800830 | 1.44 |
ENSMUST00000144602.2
|
B3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr7_+_100970910 | 1.44 |
ENSMUST00000174291.8
ENSMUST00000167888.9 ENSMUST00000172662.2 |
Stard10
|
START domain containing 10 |
| chr10_+_128745214 | 1.43 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chr5_-_44259010 | 1.42 |
ENSMUST00000087441.11
|
Prom1
|
prominin 1 |
| chr10_+_12966532 | 1.41 |
ENSMUST00000121646.8
ENSMUST00000121325.8 ENSMUST00000121766.8 |
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr9_-_7184440 | 1.41 |
ENSMUST00000140466.8
|
Dync2h1
|
dynein cytoplasmic 2 heavy chain 1 |
| chr2_-_85027041 | 1.40 |
ENSMUST00000099930.9
ENSMUST00000111601.2 |
Lrrc55
|
leucine rich repeat containing 55 |
| chr9_+_21077010 | 1.40 |
ENSMUST00000039413.15
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr13_-_94422337 | 1.35 |
ENSMUST00000022197.15
ENSMUST00000152555.8 |
Scamp1
|
secretory carrier membrane protein 1 |
| chr9_+_57604895 | 1.35 |
ENSMUST00000034865.6
|
Cyp1a1
|
cytochrome P450, family 1, subfamily a, polypeptide 1 |
| chr9_-_53521585 | 1.34 |
ENSMUST00000034547.6
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr16_+_91855158 | 1.33 |
ENSMUST00000047429.9
ENSMUST00000232677.2 ENSMUST00000113975.3 |
Mrps6
Gm49711
Slc5a3
|
mitochondrial ribosomal protein S6 predicted gene, 49711 solute carrier family 5 (inositol transporters), member 3 |
| chr8_+_106245368 | 1.31 |
ENSMUST00000034363.7
|
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr10_+_122284404 | 1.29 |
ENSMUST00000020323.7
|
Avpr1a
|
arginine vasopressin receptor 1A |
| chr1_-_23141324 | 1.27 |
ENSMUST00000073179.6
|
Ppp1r14bl
|
protein phosphatase 1, regulatory inhibitor subunit 14B like |
| chr4_-_11966367 | 1.27 |
ENSMUST00000056050.5
ENSMUST00000108299.2 ENSMUST00000108297.3 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr13_+_58956077 | 1.23 |
ENSMUST00000109838.10
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr9_-_43017249 | 1.22 |
ENSMUST00000165665.9
|
Arhgef12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr4_-_107889136 | 1.21 |
ENSMUST00000106708.8
|
Podn
|
podocan |
| chr5_+_111881790 | 1.20 |
ENSMUST00000180627.2
|
Gm26897
|
predicted gene, 26897 |
| chr13_+_104246259 | 1.17 |
ENSMUST00000160322.8
ENSMUST00000159574.2 |
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chrX_+_139565657 | 1.16 |
ENSMUST00000112990.8
ENSMUST00000112988.8 |
Mid2
|
midline 2 |
| chr1_+_21288799 | 1.14 |
ENSMUST00000027065.12
|
Tmem14a
|
transmembrane protein 14A |
| chr12_+_30961650 | 1.13 |
ENSMUST00000020997.15
ENSMUST00000110880.3 |
Sh3yl1
|
Sh3 domain YSC-like 1 |
| chr1_-_152642032 | 1.13 |
ENSMUST00000111859.8
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr19_+_22116392 | 1.12 |
ENSMUST00000237357.2
ENSMUST00000237820.2 ENSMUST00000236312.2 ENSMUST00000074770.13 ENSMUST00000087576.12 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr13_-_25454058 | 1.11 |
ENSMUST00000057866.13
|
Nrsn1
|
neurensin 1 |
| chr6_+_125169117 | 1.08 |
ENSMUST00000032485.7
|
Mrpl51
|
mitochondrial ribosomal protein L51 |
| chr18_-_68433265 | 1.08 |
ENSMUST00000152193.2
|
Fam210a
|
family with sequence similarity 210, member A |
| chr13_+_104246245 | 1.07 |
ENSMUST00000044385.14
|
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr8_-_47742429 | 1.07 |
ENSMUST00000079195.6
|
Stox2
|
storkhead box 2 |
| chr3_+_81839908 | 1.06 |
ENSMUST00000029649.3
|
Ctso
|
cathepsin O |
| chr4_-_24851086 | 1.06 |
ENSMUST00000084781.6
ENSMUST00000108218.10 |
Klhl32
|
kelch-like 32 |
| chr2_-_93164812 | 1.05 |
ENSMUST00000111265.9
|
Tspan18
|
tetraspanin 18 |
| chr3_+_40663285 | 1.04 |
ENSMUST00000091184.9
|
Slc25a31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 |
| chr6_+_29402868 | 1.02 |
ENSMUST00000154619.5
|
Ccdc136
|
coiled-coil domain containing 136 |
| chr12_+_76371634 | 1.02 |
ENSMUST00000154078.3
ENSMUST00000095610.9 |
Akap5
|
A kinase (PRKA) anchor protein 5 |
| chr3_-_131196213 | 1.02 |
ENSMUST00000197057.2
|
Sgms2
|
sphingomyelin synthase 2 |
| chr1_+_21288886 | 0.99 |
ENSMUST00000027064.8
|
Tmem14a
|
transmembrane protein 14A |
| chr5_+_93045837 | 0.98 |
ENSMUST00000113051.9
|
Shroom3
|
shroom family member 3 |
| chr8_+_105996469 | 0.95 |
ENSMUST00000172525.8
ENSMUST00000174837.8 ENSMUST00000173859.2 |
Hsf4
|
heat shock transcription factor 4 |
| chr5_-_8417982 | 0.95 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr7_+_51160855 | 0.94 |
ENSMUST00000207717.2
|
Ano5
|
anoctamin 5 |
| chr6_+_47221372 | 0.93 |
ENSMUST00000060839.8
|
Cntnap2
|
contactin associated protein-like 2 |
| chr11_+_102080489 | 0.92 |
ENSMUST00000078975.8
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr8_+_120173458 | 0.92 |
ENSMUST00000098363.10
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr14_-_30850881 | 0.88 |
ENSMUST00000203261.3
|
Smim4
|
small integral membrane protein 4 |
| chr7_-_62069887 | 0.88 |
ENSMUST00000094340.4
|
Mkrn3
|
makorin, ring finger protein, 3 |
| chr11_+_102080446 | 0.88 |
ENSMUST00000070334.10
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr11_+_118319319 | 0.86 |
ENSMUST00000017590.9
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr4_+_104224774 | 0.85 |
ENSMUST00000106830.9
|
Dab1
|
disabled 1 |
| chr7_+_49624978 | 0.83 |
ENSMUST00000107603.2
|
Nell1
|
NEL-like 1 |
| chr5_-_122187884 | 0.82 |
ENSMUST00000111752.10
|
Cux2
|
cut-like homeobox 2 |
| chr10_+_79977291 | 0.82 |
ENSMUST00000105367.8
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr4_-_108240546 | 0.81 |
ENSMUST00000053157.7
|
Shisal2a
|
shisa like 2A |
| chr12_+_76371679 | 0.81 |
ENSMUST00000172992.2
|
Akap5
|
A kinase (PRKA) anchor protein 5 |
| chr6_+_47221293 | 0.80 |
ENSMUST00000199100.5
|
Cntnap2
|
contactin associated protein-like 2 |
| chr12_+_76353835 | 0.79 |
ENSMUST00000220321.2
|
Mthfd1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
| chr11_-_97591150 | 0.79 |
ENSMUST00000018681.14
|
Pcgf2
|
polycomb group ring finger 2 |
| chr18_-_36587573 | 0.79 |
ENSMUST00000025204.7
ENSMUST00000237792.2 |
Pfdn1
|
prefoldin 1 |
| chr8_-_25164767 | 0.78 |
ENSMUST00000033957.12
|
Adam18
|
a disintegrin and metallopeptidase domain 18 |
| chr4_+_126647563 | 0.77 |
ENSMUST00000047431.11
ENSMUST00000102607.10 ENSMUST00000132660.2 |
AU040320
|
expressed sequence AU040320 |
| chr13_-_14697770 | 0.76 |
ENSMUST00000110516.3
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr7_-_44518830 | 0.76 |
ENSMUST00000208682.2
|
Ptov1
|
prostate tumor over expressed gene 1 |
| chr13_+_58955506 | 0.75 |
ENSMUST00000079828.7
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr15_+_98687720 | 0.74 |
ENSMUST00000023734.8
|
Wnt1
|
wingless-type MMTV integration site family, member 1 |
| chr2_+_127112127 | 0.74 |
ENSMUST00000110375.9
|
Stard7
|
START domain containing 7 |
| chr2_+_127112613 | 0.74 |
ENSMUST00000125049.2
ENSMUST00000110374.2 |
Stard7
|
START domain containing 7 |
| chr2_+_140237366 | 0.73 |
ENSMUST00000110061.2
|
Macrod2
|
mono-ADP ribosylhydrolase 2 |
| chr11_-_69652789 | 0.73 |
ENSMUST00000102586.5
|
Slc35g3
|
solute carrier family 35, member G3 |
| chr8_-_47742389 | 0.72 |
ENSMUST00000211737.2
|
Stox2
|
storkhead box 2 |
| chr5_+_91079068 | 0.72 |
ENSMUST00000202781.2
ENSMUST00000071652.6 |
Mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr2_-_164032521 | 0.71 |
ENSMUST00000051272.8
|
Wfdc12
|
WAP four-disulfide core domain 12 |
| chr8_+_105996419 | 0.70 |
ENSMUST00000036127.9
ENSMUST00000163734.9 |
Hsf4
|
heat shock transcription factor 4 |
| chr4_-_126647156 | 0.68 |
ENSMUST00000030637.14
ENSMUST00000106116.2 |
Ncdn
|
neurochondrin |
| chr8_+_72021567 | 0.68 |
ENSMUST00000034267.5
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr6_+_22288220 | 0.68 |
ENSMUST00000128245.8
ENSMUST00000031681.10 ENSMUST00000148639.2 |
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr5_-_25305621 | 0.68 |
ENSMUST00000030784.14
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr1_+_120268299 | 0.68 |
ENSMUST00000037286.12
|
C1ql2
|
complement component 1, q subcomponent-like 2 |
| chr8_-_88199231 | 0.66 |
ENSMUST00000034076.16
|
Cbln1
|
cerebellin 1 precursor protein |
| chr11_-_69652670 | 0.65 |
ENSMUST00000231829.2
|
Slc35g3
|
solute carrier family 35, member G3 |
| chr1_-_186875270 | 0.64 |
ENSMUST00000184543.2
|
Spata17
|
spermatogenesis associated 17 |
| chr12_+_80565764 | 0.64 |
ENSMUST00000021558.8
|
Galnt16
|
polypeptide N-acetylgalactosaminyltransferase 16 |
| chr8_-_71834543 | 0.63 |
ENSMUST00000002466.9
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr8_+_72021510 | 0.61 |
ENSMUST00000212889.2
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr17_-_26417982 | 0.61 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr1_+_90842785 | 0.60 |
ENSMUST00000027528.7
|
Mlph
|
melanophilin |
| chr5_+_125080504 | 0.60 |
ENSMUST00000197746.2
|
Rflna
|
refilin A |
| chr17_+_21165573 | 0.59 |
ENSMUST00000007708.14
|
Ppp2r1a
|
protein phosphatase 2, regulatory subunit A, alpha |
| chrX_+_68403900 | 0.58 |
ENSMUST00000033532.7
|
Aff2
|
AF4/FMR2 family, member 2 |
| chr1_-_37469037 | 0.58 |
ENSMUST00000027286.7
|
Coa5
|
cytochrome C oxidase assembly factor 5 |
| chr4_+_28813152 | 0.58 |
ENSMUST00000108194.9
ENSMUST00000108191.2 |
Epha7
|
Eph receptor A7 |
| chr7_+_79910948 | 0.56 |
ENSMUST00000117989.2
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr4_+_155875629 | 0.55 |
ENSMUST00000105593.2
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr6_-_89339581 | 0.53 |
ENSMUST00000163139.8
|
Plxna1
|
plexin A1 |
| chr10_+_17598961 | 0.52 |
ENSMUST00000038107.9
ENSMUST00000219558.2 ENSMUST00000218370.2 |
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr4_+_28813125 | 0.52 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chr1_+_164076621 | 0.51 |
ENSMUST00000159230.8
|
Slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr6_+_17306414 | 0.51 |
ENSMUST00000150901.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr7_-_34354924 | 0.49 |
ENSMUST00000032709.3
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr1_+_164076592 | 0.49 |
ENSMUST00000044021.12
|
Slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr6_+_17306334 | 0.47 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr12_-_11200306 | 0.46 |
ENSMUST00000055673.2
|
Kcns3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr10_+_127919142 | 0.46 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr12_-_27392356 | 0.45 |
ENSMUST00000079063.7
|
Sox11
|
SRY (sex determining region Y)-box 11 |
| chr15_-_38976034 | 0.45 |
ENSMUST00000227323.2
ENSMUST00000022908.10 |
Slc25a32
|
solute carrier family 25, member 32 |
| chr13_-_114595475 | 0.44 |
ENSMUST00000022287.8
ENSMUST00000223640.3 |
Fst
|
follistatin |
| chr7_+_6463510 | 0.44 |
ENSMUST00000056120.5
|
Olfr1336
|
olfactory receptor 1336 |
| chr8_-_94739469 | 0.44 |
ENSMUST00000053766.14
|
Amfr
|
autocrine motility factor receptor |
| chr1_-_30988381 | 0.43 |
ENSMUST00000232841.2
|
Ptp4a1
|
protein tyrosine phosphatase 4a1 |
| chr2_-_36995660 | 0.43 |
ENSMUST00000100144.2
|
Olfr362
|
olfactory receptor 362 |
| chr15_-_7427815 | 0.42 |
ENSMUST00000096494.5
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr1_-_120192977 | 0.41 |
ENSMUST00000140490.8
ENSMUST00000112640.8 |
Steap3
|
STEAP family member 3 |
| chr15_-_7427759 | 0.41 |
ENSMUST00000058593.10
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr15_+_97862081 | 0.41 |
ENSMUST00000229433.2
ENSMUST00000229428.2 ENSMUST00000064200.9 ENSMUST00000230072.2 ENSMUST00000231144.2 |
Tmem106c
|
transmembrane protein 106C |
| chr15_+_34838195 | 0.40 |
ENSMUST00000228725.2
|
Kcns2
|
K+ voltage-gated channel, subfamily S, 2 |
| chr11_+_58668915 | 0.40 |
ENSMUST00000081533.5
|
Olfr315
|
olfactory receptor 315 |
| chr8_+_85786684 | 0.38 |
ENSMUST00000095220.4
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr15_-_79212400 | 0.37 |
ENSMUST00000173163.8
ENSMUST00000047816.15 ENSMUST00000172403.9 ENSMUST00000173632.8 |
Pla2g6
|
phospholipase A2, group VI |
| chr11_+_69983459 | 0.36 |
ENSMUST00000102572.8
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr11_+_103857541 | 0.36 |
ENSMUST00000057921.10
ENSMUST00000063347.12 |
Arf2
|
ADP-ribosylation factor 2 |
| chr6_+_17306379 | 0.35 |
ENSMUST00000115455.3
|
Cav1
|
caveolin 1, caveolae protein |
| chr13_-_38221045 | 0.35 |
ENSMUST00000089840.5
|
Cage1
|
cancer antigen 1 |
| chr19_+_36811615 | 0.35 |
ENSMUST00000025729.12
|
Tnks2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chr9_+_39933648 | 0.33 |
ENSMUST00000059859.5
|
Olfr981
|
olfactory receptor 981 |
| chr11_+_113510207 | 0.33 |
ENSMUST00000106630.2
|
Sstr2
|
somatostatin receptor 2 |
| chr5_-_139470169 | 0.33 |
ENSMUST00000150992.2
ENSMUST00000110851.8 ENSMUST00000079996.13 |
Zfand2a
|
zinc finger, AN1-type domain 2A |
| chr3_+_40904253 | 0.30 |
ENSMUST00000048490.13
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr14_+_33041660 | 0.29 |
ENSMUST00000111955.2
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr2_-_113844100 | 0.28 |
ENSMUST00000090275.5
|
Gjd2
|
gap junction protein, delta 2 |
| chr10_-_79449826 | 0.28 |
ENSMUST00000059699.9
ENSMUST00000178228.3 |
C2cd4c
|
C2 calcium-dependent domain containing 4C |
| chr9_+_108925727 | 0.27 |
ENSMUST00000130366.2
|
Plxnb1
|
plexin B1 |
| chr17_-_28299544 | 0.27 |
ENSMUST00000042692.13
ENSMUST00000114836.9 |
Tcp11
|
t-complex protein 11 |
| chr11_+_69983479 | 0.27 |
ENSMUST00000143772.8
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr9_-_46227219 | 0.26 |
ENSMUST00000160795.2
|
4931429L15Rik
|
RIKEN cDNA 4931429L15 gene |
| chr9_+_44013019 | 0.25 |
ENSMUST00000034654.9
ENSMUST00000206308.2 ENSMUST00000161381.8 |
Mfrp
|
membrane frizzled-related protein |
| chr10_-_112764879 | 0.25 |
ENSMUST00000099276.4
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr11_+_69983531 | 0.25 |
ENSMUST00000124721.2
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr13_+_58955675 | 0.25 |
ENSMUST00000224402.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr13_-_14697681 | 0.24 |
ENSMUST00000223189.2
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr19_-_6910922 | 0.24 |
ENSMUST00000235248.2
|
Kcnk4
|
potassium channel, subfamily K, member 4 |
| chr17_-_24937025 | 0.23 |
ENSMUST00000008626.10
|
Rnf151
|
ring finger protein 151 |
| chr17_-_13128768 | 0.21 |
ENSMUST00000232982.2
|
Pnldc1
|
poly(A)-specific ribonuclease (PARN)-like domain containing 1 |
| chr14_+_47120311 | 0.21 |
ENSMUST00000022386.15
ENSMUST00000228404.2 ENSMUST00000100672.11 |
Samd4
|
sterile alpha motif domain containing 4 |
| chr10_-_62486948 | 0.20 |
ENSMUST00000020270.6
|
Ddx50
|
DExD box helicase 50 |
| chr5_+_100994230 | 0.20 |
ENSMUST00000092990.4
ENSMUST00000145612.2 |
Gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr17_-_88105422 | 0.19 |
ENSMUST00000055221.9
|
Kcnk12
|
potassium channel, subfamily K, member 12 |
| chr17_-_33435325 | 0.18 |
ENSMUST00000112162.4
|
Olfr1564
|
olfactory receptor 1564 |
| chr17_-_28299569 | 0.17 |
ENSMUST00000129046.9
ENSMUST00000043925.16 |
Tcp11
|
t-complex protein 11 |
| chr4_+_148888877 | 0.16 |
ENSMUST00000094464.10
ENSMUST00000122222.8 |
Casz1
|
castor zinc finger 1 |
| chr15_-_79212323 | 0.16 |
ENSMUST00000166977.9
|
Pla2g6
|
phospholipase A2, group VI |
| chr11_+_101046708 | 0.15 |
ENSMUST00000043654.10
|
Tubg2
|
tubulin, gamma 2 |
| chr16_-_4442802 | 0.15 |
ENSMUST00000014445.7
|
Pam16
|
presequence translocase-asssociated motor 16 homolog (S. cerevisiae) |
| chr10_-_10956700 | 0.14 |
ENSMUST00000105560.2
|
Grm1
|
glutamate receptor, metabotropic 1 |
| chr11_+_113510135 | 0.14 |
ENSMUST00000146390.3
|
Sstr2
|
somatostatin receptor 2 |
| chr8_+_120339440 | 0.14 |
ENSMUST00000098361.4
|
Adad2
|
adenosine deaminase domain containing 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.9 | 2.8 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.7 | 2.0 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.7 | 2.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.6 | 9.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.6 | 1.9 | GO:0019405 | hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) |
| 0.6 | 4.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.6 | 5.7 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.5 | 2.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.5 | 5.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.4 | 1.3 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.4 | 1.3 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.4 | 1.3 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.4 | 1.7 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.4 | 1.5 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.4 | 4.5 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.4 | 0.7 | GO:0061184 | positive regulation of dermatome development(GO:0061184) |
| 0.4 | 2.2 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.3 | 1.3 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.3 | 1.3 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.3 | 0.9 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.3 | 1.8 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.3 | 2.1 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.3 | 1.4 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.3 | 0.8 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 | 1.7 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 1.4 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.2 | 1.3 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.2 | 0.9 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 1.0 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.2 | 0.6 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 2.0 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 3.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 1.8 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 3.8 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.2 | 2.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.2 | 4.6 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.2 | 5.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 0.6 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.7 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 3.0 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 2.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 1.3 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 0.5 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.4 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.1 | 2.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 2.1 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.1 | 1.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 0.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.5 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of arachidonic acid secretion(GO:0090238) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 1.5 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 1.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.4 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.1 | 1.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.2 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.1 | 1.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 1.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.5 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.5 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 1.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 0.2 | GO:1902870 | camera-type eye photoreceptor cell fate commitment(GO:0060220) negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 0.1 | 0.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 1.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 5.3 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 1.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.7 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.8 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.5 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.0 | 1.1 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.5 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.6 | GO:0050965 | entrainment of circadian clock by photoperiod(GO:0043153) detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.9 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 1.8 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 1.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.7 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.6 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.0 | 0.4 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.0 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 1.0 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 9.6 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 1.3 | 3.8 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.5 | 3.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.5 | 2.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.5 | 1.4 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.3 | 5.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.3 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 2.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 1.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 2.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 6.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 7.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.5 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 3.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 5.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 1.1 | 3.2 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.7 | 2.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.6 | 3.8 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.5 | 1.8 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.4 | 1.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.4 | 2.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.4 | 4.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 2.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.3 | 2.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.3 | 2.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 5.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 1.3 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.3 | 1.5 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.2 | 1.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.2 | 1.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 0.9 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.2 | 1.3 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.2 | 1.3 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.2 | 9.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 3.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 1.0 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 2.8 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 2.4 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 2.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.3 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.1 | 1.8 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 1.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) axon guidance receptor activity(GO:0008046) |
| 0.1 | 2.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 1.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 2.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.2 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.1 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 1.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 2.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 4.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 4.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.4 | GO:0052851 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.6 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.5 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 1.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 2.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 9.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 4.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 3.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 2.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 4.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 4.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 9.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 3.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 2.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.8 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.0 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 1.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |