Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
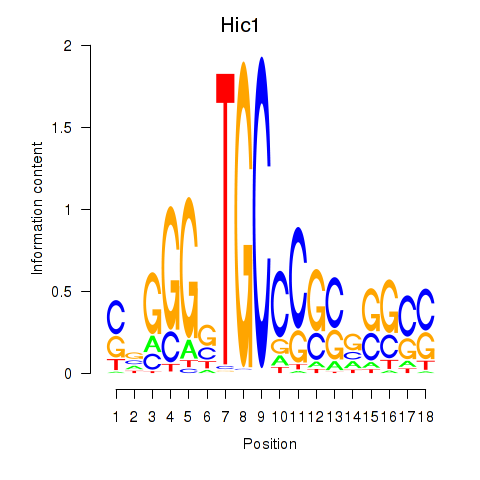
Results for Hic1
Z-value: 1.83
Transcription factors associated with Hic1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hic1
|
ENSMUSG00000043099.5 | Hic1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hic1 | mm39_v1_chr11_-_75060345_75060345 | 0.05 | 6.9e-01 | Click! |
Activity profile of Hic1 motif
Sorted Z-values of Hic1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hic1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_121279062 | 11.39 |
ENSMUST00000106107.3
|
Rab40b
|
Rab40B, member RAS oncogene family |
| chr12_-_87037204 | 10.59 |
ENSMUST00000222543.2
|
Zdhhc22
|
zinc finger, DHHC-type containing 22 |
| chr4_-_117740624 | 10.52 |
ENSMUST00000030266.12
|
B4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr12_-_109019507 | 9.97 |
ENSMUST00000185745.2
ENSMUST00000239108.2 |
Begain
|
brain-enriched guanylate kinase-associated |
| chr6_-_113172340 | 9.65 |
ENSMUST00000162280.2
|
Lhfpl4
|
lipoma HMGIC fusion partner-like protein 4 |
| chr5_+_135052336 | 9.63 |
ENSMUST00000005509.11
ENSMUST00000201008.4 |
Stx1a
|
syntaxin 1A (brain) |
| chr3_-_80820835 | 9.48 |
ENSMUST00000107743.8
ENSMUST00000029654.15 |
Glrb
|
glycine receptor, beta subunit |
| chr2_+_148237258 | 9.33 |
ENSMUST00000109962.4
|
Sstr4
|
somatostatin receptor 4 |
| chr4_+_129878890 | 9.31 |
ENSMUST00000106017.8
ENSMUST00000121049.8 |
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr15_-_75438457 | 9.26 |
ENSMUST00000163116.8
ENSMUST00000023241.12 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr5_-_52628825 | 9.10 |
ENSMUST00000198008.5
ENSMUST00000059428.7 |
Ccdc149
|
coiled-coil domain containing 149 |
| chr4_-_124755964 | 9.06 |
ENSMUST00000064444.8
|
Maneal
|
mannosidase, endo-alpha-like |
| chr5_+_9316097 | 8.85 |
ENSMUST00000134991.8
ENSMUST00000069538.14 ENSMUST00000115348.9 |
Elapor2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr10_-_9550907 | 8.68 |
ENSMUST00000100070.5
|
Samd5
|
sterile alpha motif domain containing 5 |
| chr6_+_38639945 | 8.31 |
ENSMUST00000114874.5
|
Clec2l
|
C-type lectin domain family 2, member L |
| chr9_+_26645024 | 8.28 |
ENSMUST00000160899.8
ENSMUST00000161431.3 ENSMUST00000159799.8 |
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr17_+_26028059 | 8.25 |
ENSMUST00000045692.9
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr7_-_30826376 | 8.04 |
ENSMUST00000098548.8
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr5_-_115332343 | 7.99 |
ENSMUST00000112113.8
|
Cabp1
|
calcium binding protein 1 |
| chr2_+_116951855 | 7.95 |
ENSMUST00000028829.13
|
Spred1
|
sprouty protein with EVH-1 domain 1, related sequence |
| chr5_-_71253107 | 7.85 |
ENSMUST00000197284.5
|
Gabra2
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 |
| chr2_-_73605387 | 7.82 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr13_+_55097200 | 7.81 |
ENSMUST00000026994.14
ENSMUST00000109994.9 |
Unc5a
|
unc-5 netrin receptor A |
| chr1_+_182591425 | 7.56 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr19_+_47003111 | 7.40 |
ENSMUST00000037636.4
|
Ina
|
internexin neuronal intermediate filament protein, alpha |
| chr1_-_33946802 | 7.32 |
ENSMUST00000115161.8
ENSMUST00000129464.8 ENSMUST00000062289.11 |
Bend6
|
BEN domain containing 6 |
| chr8_+_123844090 | 7.31 |
ENSMUST00000037900.9
|
Cpne7
|
copine VII |
| chr12_+_74044435 | 7.27 |
ENSMUST00000221220.2
|
Syt16
|
synaptotagmin XVI |
| chr11_-_102338473 | 7.26 |
ENSMUST00000049057.5
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr9_-_56703422 | 7.26 |
ENSMUST00000210032.2
|
Lingo1
|
leucine rich repeat and Ig domain containing 1 |
| chr9_-_66951234 | 7.10 |
ENSMUST00000113690.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr19_+_47167259 | 6.96 |
ENSMUST00000111808.11
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr11_-_6015538 | 6.94 |
ENSMUST00000101585.10
ENSMUST00000066431.14 ENSMUST00000109815.9 ENSMUST00000109812.9 ENSMUST00000101586.3 ENSMUST00000093355.12 ENSMUST00000019133.11 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta |
| chr12_-_109034099 | 6.61 |
ENSMUST00000190647.3
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr2_-_32737238 | 6.52 |
ENSMUST00000050000.16
|
Stxbp1
|
syntaxin binding protein 1 |
| chrX_-_134968985 | 6.48 |
ENSMUST00000049130.8
|
Bex2
|
brain expressed X-linked 2 |
| chr7_-_84059321 | 6.42 |
ENSMUST00000085077.5
ENSMUST00000207769.2 |
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr5_+_16139760 | 6.40 |
ENSMUST00000101581.10
ENSMUST00000039370.14 ENSMUST00000199704.5 ENSMUST00000180204.8 ENSMUST00000078272.13 ENSMUST00000115281.7 |
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr12_-_81379903 | 6.40 |
ENSMUST00000085238.13
ENSMUST00000182208.8 |
Slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr16_-_20060053 | 6.39 |
ENSMUST00000040880.9
|
Map6d1
|
MAP6 domain containing 1 |
| chr10_-_43050516 | 6.37 |
ENSMUST00000040275.9
|
Sobp
|
sine oculis binding protein |
| chr5_-_31453206 | 6.37 |
ENSMUST00000041266.11
ENSMUST00000172435.8 ENSMUST00000201417.2 |
Fndc4
|
fibronectin type III domain containing 4 |
| chr4_+_138181616 | 6.36 |
ENSMUST00000050918.4
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr2_+_28095660 | 6.31 |
ENSMUST00000102879.4
ENSMUST00000028177.11 |
Olfm1
|
olfactomedin 1 |
| chr6_+_71684846 | 6.29 |
ENSMUST00000212792.2
|
Reep1
|
receptor accessory protein 1 |
| chr13_+_104246245 | 6.29 |
ENSMUST00000044385.14
|
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr6_+_21215472 | 6.25 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chrX_+_10351360 | 6.24 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr5_-_112542671 | 6.21 |
ENSMUST00000196256.2
|
Asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr13_+_58954374 | 6.21 |
ENSMUST00000225488.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr13_+_104246259 | 6.17 |
ENSMUST00000160322.8
ENSMUST00000159574.2 |
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr11_-_28534260 | 6.11 |
ENSMUST00000093253.10
ENSMUST00000109502.9 ENSMUST00000042534.15 |
Ccdc85a
|
coiled-coil domain containing 85A |
| chr4_+_33924632 | 6.10 |
ENSMUST00000057188.7
|
Cnr1
|
cannabinoid receptor 1 (brain) |
| chr13_+_58954447 | 6.04 |
ENSMUST00000224259.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr15_+_89407954 | 5.98 |
ENSMUST00000230807.2
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr10_-_77254041 | 5.97 |
ENSMUST00000020496.14
ENSMUST00000098374.9 ENSMUST00000105406.8 |
Adarb1
|
adenosine deaminase, RNA-specific, B1 |
| chr2_-_73605684 | 5.93 |
ENSMUST00000112024.10
ENSMUST00000180045.8 |
Chn1
|
chimerin 1 |
| chr2_+_24944407 | 5.87 |
ENSMUST00000102931.11
ENSMUST00000074422.14 ENSMUST00000132172.8 ENSMUST00000114388.8 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr6_+_71684872 | 5.85 |
ENSMUST00000212631.2
|
Reep1
|
receptor accessory protein 1 |
| chr5_+_16139683 | 5.77 |
ENSMUST00000167946.9
|
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr4_-_141966662 | 5.75 |
ENSMUST00000036476.10
|
Kazn
|
kazrin, periplakin interacting protein |
| chr7_-_81104423 | 5.73 |
ENSMUST00000178892.3
ENSMUST00000098331.10 |
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr5_+_33879018 | 5.69 |
ENSMUST00000201437.4
ENSMUST00000067150.14 ENSMUST00000169212.9 ENSMUST00000114411.9 ENSMUST00000164207.10 ENSMUST00000087820.8 |
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr4_-_81360881 | 5.66 |
ENSMUST00000220807.2
|
Mpdz
|
multiple PDZ domain crumbs cell polarity complex component |
| chr4_-_81360993 | 5.62 |
ENSMUST00000107262.8
ENSMUST00000102830.10 |
Mpdz
|
multiple PDZ domain crumbs cell polarity complex component |
| chr17_+_25936463 | 5.59 |
ENSMUST00000115108.4
|
Gng13
|
guanine nucleotide binding protein (G protein), gamma 13 |
| chr5_+_113638313 | 5.59 |
ENSMUST00000094452.4
|
Wscd2
|
WSC domain containing 2 |
| chr2_-_24653059 | 5.57 |
ENSMUST00000100348.10
ENSMUST00000041342.12 ENSMUST00000114447.8 ENSMUST00000102939.9 ENSMUST00000070864.14 |
Cacna1b
|
calcium channel, voltage-dependent, N type, alpha 1B subunit |
| chr7_-_45016138 | 5.55 |
ENSMUST00000211067.2
ENSMUST00000003961.16 |
Ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr2_+_24944367 | 5.54 |
ENSMUST00000100334.11
ENSMUST00000152122.8 ENSMUST00000116574.10 ENSMUST00000006646.15 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr8_+_70768409 | 5.53 |
ENSMUST00000165819.9
ENSMUST00000140239.4 |
Cers1
|
ceramide synthase 1 |
| chr2_+_119629995 | 5.45 |
ENSMUST00000028763.10
|
Tyro3
|
TYRO3 protein tyrosine kinase 3 |
| chr7_+_46045862 | 5.43 |
ENSMUST00000025202.8
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1 |
| chr11_+_49792627 | 5.40 |
ENSMUST00000093141.12
ENSMUST00000093142.12 |
Rasgef1c
|
RasGEF domain family, member 1C |
| chr7_-_57159119 | 5.40 |
ENSMUST00000206382.2
|
Gabra5
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5 |
| chr2_+_164327988 | 5.39 |
ENSMUST00000109350.9
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr1_-_79836344 | 5.39 |
ENSMUST00000027467.11
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr1_-_25868788 | 5.35 |
ENSMUST00000151309.8
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr2_-_26012751 | 5.34 |
ENSMUST00000140993.2
ENSMUST00000028300.6 |
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr12_+_37930661 | 5.34 |
ENSMUST00000040500.9
|
Dgkb
|
diacylglycerol kinase, beta |
| chr8_-_86427383 | 5.28 |
ENSMUST00000209479.2
|
Neto2
|
neuropilin (NRP) and tolloid (TLL)-like 2 |
| chr2_+_28083105 | 5.27 |
ENSMUST00000100244.10
|
Olfm1
|
olfactomedin 1 |
| chrX_+_70093766 | 5.27 |
ENSMUST00000239162.2
ENSMUST00000114629.4 ENSMUST00000082088.10 |
Mamld1
|
mastermind-like domain containing 1 |
| chr7_-_105282687 | 5.24 |
ENSMUST00000147044.4
ENSMUST00000106791.8 ENSMUST00000153371.9 ENSMUST00000106789.8 |
Trim3
|
tripartite motif-containing 3 |
| chr17_+_70276382 | 5.23 |
ENSMUST00000146730.9
|
Dlgap1
|
DLG associated protein 1 |
| chr13_+_74157538 | 5.23 |
ENSMUST00000022057.9
|
Tppp
|
tubulin polymerization promoting protein |
| chr11_-_69496655 | 5.23 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr2_+_90716204 | 5.22 |
ENSMUST00000111466.3
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4 |
| chr10_+_11219117 | 5.22 |
ENSMUST00000069106.5
|
Epm2a
|
epilepsy, progressive myoclonic epilepsy, type 2 gene alpha |
| chr3_+_146558114 | 5.20 |
ENSMUST00000170055.8
ENSMUST00000037942.11 |
Ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr1_+_63485332 | 5.19 |
ENSMUST00000114103.8
ENSMUST00000114107.3 |
Adam23
|
a disintegrin and metallopeptidase domain 23 |
| chr2_+_24944480 | 5.14 |
ENSMUST00000114386.8
|
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr19_-_5148506 | 5.12 |
ENSMUST00000025805.8
|
Cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr4_-_46991842 | 5.10 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr10_-_79473675 | 5.09 |
ENSMUST00000020564.7
|
Shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr14_-_78970160 | 5.07 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr9_+_26645141 | 5.07 |
ENSMUST00000115269.9
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr5_-_99185201 | 5.06 |
ENSMUST00000161490.8
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr7_+_3381434 | 5.04 |
ENSMUST00000092891.6
|
Cacng7
|
calcium channel, voltage-dependent, gamma subunit 7 |
| chr13_-_69887964 | 5.00 |
ENSMUST00000065118.7
|
Ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr19_+_10366753 | 4.93 |
ENSMUST00000169121.9
ENSMUST00000076968.11 ENSMUST00000235479.2 ENSMUST00000223586.2 ENSMUST00000235784.2 ENSMUST00000224135.3 ENSMUST00000225452.3 ENSMUST00000237366.2 |
Syt7
|
synaptotagmin VII |
| chr14_-_24054927 | 4.91 |
ENSMUST00000145596.3
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr8_-_85526653 | 4.86 |
ENSMUST00000126806.2
ENSMUST00000076715.13 |
Nfix
|
nuclear factor I/X |
| chr8_+_96404713 | 4.82 |
ENSMUST00000041318.14
|
Ndrg4
|
N-myc downstream regulated gene 4 |
| chr2_-_104240679 | 4.82 |
ENSMUST00000136156.9
ENSMUST00000141159.9 ENSMUST00000089726.10 |
D430041D05Rik
|
RIKEN cDNA D430041D05 gene |
| chr4_-_138820269 | 4.82 |
ENSMUST00000042844.7
|
Nbl1
|
NBL1, DAN family BMP antagonist |
| chr12_+_37930305 | 4.79 |
ENSMUST00000220990.2
|
Dgkb
|
diacylglycerol kinase, beta |
| chr6_-_114018982 | 4.78 |
ENSMUST00000101045.10
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr2_-_32737208 | 4.78 |
ENSMUST00000077458.7
ENSMUST00000208840.2 |
Stxbp1
|
syntaxin binding protein 1 |
| chr16_-_60425608 | 4.75 |
ENSMUST00000068860.13
|
Epha6
|
Eph receptor A6 |
| chr5_+_37025810 | 4.73 |
ENSMUST00000031003.11
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr2_-_151474391 | 4.72 |
ENSMUST00000137936.2
ENSMUST00000146172.8 ENSMUST00000094456.10 ENSMUST00000148755.8 ENSMUST00000109875.8 ENSMUST00000028951.14 ENSMUST00000109877.10 |
Snph
|
syntaphilin |
| chr9_+_58395850 | 4.72 |
ENSMUST00000085658.5
|
Insyn1
|
inhibitory synaptic factor 1 |
| chr7_-_30826184 | 4.66 |
ENSMUST00000211945.2
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr2_+_31135813 | 4.66 |
ENSMUST00000000199.8
|
Ncs1
|
neuronal calcium sensor 1 |
| chr8_-_125161061 | 4.64 |
ENSMUST00000140012.8
|
Pgbd5
|
piggyBac transposable element derived 5 |
| chr1_-_134163102 | 4.63 |
ENSMUST00000187631.2
ENSMUST00000038191.8 ENSMUST00000086465.6 |
Adora1
|
adenosine A1 receptor |
| chr3_+_7568481 | 4.62 |
ENSMUST00000051064.9
ENSMUST00000193010.2 |
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr11_-_72026547 | 4.60 |
ENSMUST00000108508.3
ENSMUST00000075258.13 |
Pitpnm3
|
PITPNM family member 3 |
| chr5_+_146321757 | 4.59 |
ENSMUST00000016143.9
|
Wasf3
|
WASP family, member 3 |
| chr11_-_75686874 | 4.57 |
ENSMUST00000021209.8
|
Doc2b
|
double C2, beta |
| chr6_-_148345834 | 4.52 |
ENSMUST00000060095.15
ENSMUST00000100772.10 |
Tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr12_+_80565764 | 4.52 |
ENSMUST00000021558.8
|
Galnt16
|
polypeptide N-acetylgalactosaminyltransferase 16 |
| chr2_+_24944457 | 4.51 |
ENSMUST00000140737.8
|
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr15_-_75439013 | 4.50 |
ENSMUST00000156032.2
ENSMUST00000127095.8 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr6_+_30738043 | 4.47 |
ENSMUST00000163949.9
ENSMUST00000124665.3 |
Mest
|
mesoderm specific transcript |
| chr3_+_54662995 | 4.46 |
ENSMUST00000029371.3
|
Smad9
|
SMAD family member 9 |
| chr17_-_24424456 | 4.46 |
ENSMUST00000201583.2
ENSMUST00000202925.4 ENSMUST00000167791.9 ENSMUST00000201960.4 ENSMUST00000040474.11 ENSMUST00000201089.4 ENSMUST00000201301.4 ENSMUST00000201805.4 ENSMUST00000168410.9 ENSMUST00000097376.10 |
Tbc1d24
|
TBC1 domain family, member 24 |
| chr6_+_117988399 | 4.45 |
ENSMUST00000164960.4
|
Rasgef1a
|
RasGEF domain family, member 1A |
| chr9_-_108474757 | 4.41 |
ENSMUST00000193621.2
ENSMUST00000006853.11 |
P4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr10_-_33500583 | 4.40 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr11_-_101917745 | 4.36 |
ENSMUST00000107167.2
ENSMUST00000062801.11 |
Mpp3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
| chr8_-_68514879 | 4.34 |
ENSMUST00000212505.2
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr6_+_22875494 | 4.34 |
ENSMUST00000090568.7
|
Ptprz1
|
protein tyrosine phosphatase, receptor type Z, polypeptide 1 |
| chr18_+_82493284 | 4.33 |
ENSMUST00000047865.14
|
Mbp
|
myelin basic protein |
| chr5_-_52723607 | 4.33 |
ENSMUST00000199942.5
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr15_-_79688910 | 4.32 |
ENSMUST00000175858.10
ENSMUST00000023057.10 |
Nptxr
|
neuronal pentraxin receptor |
| chr6_+_79995860 | 4.31 |
ENSMUST00000147663.8
ENSMUST00000128718.8 ENSMUST00000126005.8 ENSMUST00000133918.8 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr7_-_84059170 | 4.30 |
ENSMUST00000208995.2
|
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr10_-_80679859 | 4.27 |
ENSMUST00000053986.9
|
Lingo3
|
leucine rich repeat and Ig domain containing 3 |
| chr9_+_111141164 | 4.27 |
ENSMUST00000197049.2
|
Trank1
|
tetratricopeptide repeat and ankyrin repeat containing 1 |
| chr11_+_67345895 | 4.27 |
ENSMUST00000108681.9
|
Gas7
|
growth arrest specific 7 |
| chr3_-_116762617 | 4.27 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr2_-_131987008 | 4.26 |
ENSMUST00000028815.15
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr12_-_103208402 | 4.26 |
ENSMUST00000074416.10
|
Prima1
|
proline rich membrane anchor 1 |
| chr17_-_31856109 | 4.25 |
ENSMUST00000078509.12
ENSMUST00000236853.2 ENSMUST00000067801.14 ENSMUST00000118504.9 |
Cbs
|
cystathionine beta-synthase |
| chr5_-_99184894 | 4.25 |
ENSMUST00000031277.7
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr3_+_108191398 | 4.25 |
ENSMUST00000135636.6
ENSMUST00000102632.7 |
Sort1
|
sortilin 1 |
| chr10_+_12966532 | 4.24 |
ENSMUST00000121646.8
ENSMUST00000121325.8 ENSMUST00000121766.8 |
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr10_-_80861239 | 4.21 |
ENSMUST00000055125.5
|
Diras1
|
DIRAS family, GTP-binding RAS-like 1 |
| chr13_-_78347876 | 4.20 |
ENSMUST00000091458.13
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr11_-_28533995 | 4.14 |
ENSMUST00000146385.9
|
Ccdc85a
|
coiled-coil domain containing 85A |
| chr4_-_151946124 | 4.12 |
ENSMUST00000169423.9
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr1_+_75427343 | 4.12 |
ENSMUST00000037708.10
|
Asic4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr7_-_45750153 | 4.12 |
ENSMUST00000180081.3
|
Kcnj11
|
potassium inwardly rectifying channel, subfamily J, member 11 |
| chrX_+_165021919 | 4.11 |
ENSMUST00000060210.14
ENSMUST00000112233.8 |
Gpm6b
|
glycoprotein m6b |
| chr1_-_37758863 | 4.11 |
ENSMUST00000160589.2
|
Cracdl
|
capping protein inhibiting regulator of actin like |
| chr9_-_42855775 | 4.10 |
ENSMUST00000114865.8
|
Grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr7_+_64151435 | 4.07 |
ENSMUST00000032732.15
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr4_-_151946155 | 4.05 |
ENSMUST00000049790.14
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr9_-_53975629 | 4.03 |
ENSMUST00000098760.5
|
Tnfaip8l3
|
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chr15_-_79389442 | 3.97 |
ENSMUST00000057801.8
|
Kcnj4
|
potassium inwardly-rectifying channel, subfamily J, member 4 |
| chr7_-_105282751 | 3.97 |
ENSMUST00000057525.14
|
Trim3
|
tripartite motif-containing 3 |
| chr2_-_57014015 | 3.97 |
ENSMUST00000112629.8
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr4_-_131565542 | 3.96 |
ENSMUST00000030741.9
ENSMUST00000105987.9 |
Ptpru
|
protein tyrosine phosphatase, receptor type, U |
| chr10_-_79975181 | 3.96 |
ENSMUST00000105369.8
|
Cbarp
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein |
| chr12_+_37930169 | 3.96 |
ENSMUST00000221176.2
|
Dgkb
|
diacylglycerol kinase, beta |
| chr9_+_102988940 | 3.96 |
ENSMUST00000189134.2
ENSMUST00000035155.8 |
Rab6b
|
RAB6B, member RAS oncogene family |
| chr2_-_167473892 | 3.95 |
ENSMUST00000060645.13
ENSMUST00000140216.2 ENSMUST00000151365.8 ENSMUST00000109207.10 |
Ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr18_+_77273510 | 3.95 |
ENSMUST00000075290.8
ENSMUST00000079618.11 |
St8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr11_+_17109263 | 3.95 |
ENSMUST00000102880.5
|
Ppp3r1
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) |
| chr7_-_142213219 | 3.94 |
ENSMUST00000121128.8
|
Igf2
|
insulin-like growth factor 2 |
| chr7_-_57159743 | 3.94 |
ENSMUST00000068456.8
ENSMUST00000206734.2 |
Gabra5
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5 |
| chr9_-_42175522 | 3.93 |
ENSMUST00000217513.2
ENSMUST00000052725.15 |
Sc5d
|
sterol-C5-desaturase |
| chr5_-_146521629 | 3.92 |
ENSMUST00000200112.2
ENSMUST00000197431.2 ENSMUST00000197825.2 |
Gpr12
|
G-protein coupled receptor 12 |
| chr1_+_34789662 | 3.91 |
ENSMUST00000159021.9
|
Arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr6_+_114108190 | 3.91 |
ENSMUST00000032451.9
|
Slc6a11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11 |
| chr2_+_102488985 | 3.89 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr8_+_110806390 | 3.88 |
ENSMUST00000212754.2
ENSMUST00000058804.10 |
Zfp612
|
zinc finger protein 612 |
| chr15_-_75438660 | 3.87 |
ENSMUST00000065417.15
|
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr5_+_25964985 | 3.87 |
ENSMUST00000128727.8
ENSMUST00000088244.6 |
Actr3b
|
ARP3 actin-related protein 3B |
| chr7_+_98917542 | 3.86 |
ENSMUST00000107100.3
ENSMUST00000208605.2 |
Map6
|
microtubule-associated protein 6 |
| chr12_-_76869510 | 3.85 |
ENSMUST00000154765.8
|
Rab15
|
RAB15, member RAS oncogene family |
| chr10_+_90665270 | 3.85 |
ENSMUST00000182202.8
ENSMUST00000182966.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr9_-_44646487 | 3.83 |
ENSMUST00000034611.15
|
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr13_-_47168286 | 3.83 |
ENSMUST00000052747.4
|
Nhlrc1
|
NHL repeat containing 1 |
| chr10_+_59942274 | 3.80 |
ENSMUST00000165024.3
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr6_-_119307685 | 3.78 |
ENSMUST00000112756.8
ENSMUST00000068351.14 |
Lrtm2
|
leucine-rich repeats and transmembrane domains 2 |
| chr17_+_13980764 | 3.77 |
ENSMUST00000139347.8
ENSMUST00000156591.8 ENSMUST00000170827.9 ENSMUST00000139666.8 ENSMUST00000137708.8 ENSMUST00000137784.8 ENSMUST00000150848.8 |
Afdn
|
afadin, adherens junction formation factor |
| chr6_-_126717590 | 3.76 |
ENSMUST00000185333.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chr15_-_44651411 | 3.76 |
ENSMUST00000090057.6
ENSMUST00000110269.8 ENSMUST00000228639.2 |
Sybu
|
syntabulin (syntaxin-interacting) |
| chr17_+_44112679 | 3.70 |
ENSMUST00000229744.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr13_-_10410857 | 3.70 |
ENSMUST00000187510.7
|
Chrm3
|
cholinergic receptor, muscarinic 3, cardiac |
| chr18_-_34506366 | 3.67 |
ENSMUST00000006027.7
|
Reep5
|
receptor accessory protein 5 |
| chr9_+_45341589 | 3.66 |
ENSMUST00000239471.2
ENSMUST00000034592.11 ENSMUST00000239429.2 |
Dscaml1
|
DS cell adhesion molecule like 1 |
| chr5_-_128510108 | 3.65 |
ENSMUST00000044441.8
|
Tmem132d
|
transmembrane protein 132D |
| chr13_-_99653045 | 3.64 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr2_+_156455583 | 3.63 |
ENSMUST00000109567.10
ENSMUST00000169464.9 |
Dlgap4
|
DLG associated protein 4 |
| chr10_+_20828446 | 3.62 |
ENSMUST00000105525.12
|
Ahi1
|
Abelson helper integration site 1 |
| chr8_+_127790772 | 3.62 |
ENSMUST00000079777.12
ENSMUST00000160272.8 ENSMUST00000162907.8 ENSMUST00000162536.8 ENSMUST00000026921.13 ENSMUST00000162665.8 ENSMUST00000162602.8 ENSMUST00000160581.8 ENSMUST00000161355.8 ENSMUST00000162531.8 ENSMUST00000160766.8 ENSMUST00000159537.8 |
Pard3
|
par-3 family cell polarity regulator |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.7 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 3.5 | 21.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 3.2 | 9.6 | GO:0009629 | response to gravity(GO:0009629) |
| 2.7 | 8.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 2.5 | 7.6 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 2.5 | 7.5 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 2.3 | 9.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 2.3 | 9.3 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 2.3 | 6.9 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 2.3 | 9.1 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 2.2 | 11.2 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 2.1 | 6.4 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 2.1 | 12.7 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 2.1 | 2.1 | GO:1902996 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 2.1 | 6.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 2.0 | 12.2 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 2.0 | 6.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 2.0 | 7.9 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 1.9 | 5.8 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 1.9 | 5.7 | GO:0060365 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) |
| 1.9 | 5.7 | GO:0021762 | substantia nigra development(GO:0021762) |
| 1.9 | 11.3 | GO:1903296 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 1.8 | 5.5 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 1.8 | 9.0 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 1.8 | 5.4 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 1.8 | 5.3 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 1.7 | 3.5 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 1.6 | 4.8 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 1.5 | 4.6 | GO:0042323 | positive regulation of nucleobase-containing compound transport(GO:0032241) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.4 | 4.3 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 1.4 | 18.2 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 1.4 | 7.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 1.3 | 11.9 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 1.3 | 4.0 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 1.3 | 6.5 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 1.3 | 5.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 1.3 | 6.3 | GO:0002265 | astrocyte activation involved in immune response(GO:0002265) |
| 1.2 | 4.9 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.2 | 8.6 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 1.2 | 3.6 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 1.2 | 2.4 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 1.2 | 7.0 | GO:0034465 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 1.2 | 13.9 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 1.1 | 3.4 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 1.1 | 5.5 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 1.1 | 4.3 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 1.1 | 11.8 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 1.0 | 15.5 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.0 | 7.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.0 | 4.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 1.0 | 3.0 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 1.0 | 4.8 | GO:0015888 | thiamine transport(GO:0015888) |
| 1.0 | 2.9 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 1.0 | 10.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 1.0 | 4.8 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.9 | 1.8 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.9 | 2.7 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.9 | 3.6 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.9 | 3.6 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.9 | 2.7 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.9 | 3.6 | GO:1902045 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.8 | 5.8 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.8 | 5.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.8 | 3.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.8 | 2.3 | GO:0003127 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.8 | 2.3 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.8 | 3.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.8 | 2.3 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.8 | 7.6 | GO:0046959 | habituation(GO:0046959) |
| 0.7 | 3.7 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.7 | 2.2 | GO:2000313 | regulation of cytokine activity(GO:0060300) fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.7 | 4.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.7 | 5.6 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.7 | 2.8 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.7 | 3.4 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.7 | 1.4 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.7 | 7.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.7 | 2.7 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.7 | 4.7 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.7 | 18.2 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.7 | 6.0 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.7 | 2.0 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.7 | 29.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.6 | 1.9 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.6 | 6.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.6 | 1.9 | GO:1903400 | mitochondrial ornithine transport(GO:0000066) L-ornithine transmembrane transport(GO:1903352) L-arginine transmembrane transport(GO:1903400) L-lysine transmembrane transport(GO:1903401) arginine transmembrane transport(GO:1903826) |
| 0.6 | 3.8 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.6 | 2.5 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.6 | 1.9 | GO:0006169 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 0.6 | 6.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.6 | 1.8 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.6 | 2.4 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.6 | 4.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.6 | 4.8 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.6 | 3.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.6 | 2.9 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.6 | 1.7 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.6 | 1.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.6 | 12.1 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.6 | 2.3 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.6 | 1.7 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.6 | 16.8 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.5 | 0.5 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.5 | 9.3 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.5 | 2.7 | GO:0097195 | pilomotor reflex(GO:0097195) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.5 | 4.3 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.5 | 1.6 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.5 | 2.7 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.5 | 2.7 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.5 | 19.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.5 | 1.6 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.5 | 4.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.5 | 6.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.5 | 3.1 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.5 | 4.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.5 | 1.5 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.5 | 1.5 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.5 | 1.5 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.5 | 1.5 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.5 | 1.9 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.5 | 1.4 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.5 | 4.8 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.5 | 1.9 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.5 | 3.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.5 | 3.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.5 | 2.8 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.5 | 1.9 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.5 | 1.8 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.5 | 7.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.5 | 1.4 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.5 | 0.9 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.5 | 9.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.4 | 3.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.4 | 1.3 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.4 | 0.9 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.4 | 3.8 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.4 | 2.9 | GO:0003383 | apical constriction(GO:0003383) |
| 0.4 | 2.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.4 | 1.3 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.4 | 0.8 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.4 | 3.7 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.4 | 9.3 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 0.4 | 2.8 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.4 | 5.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.4 | 2.4 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.4 | 5.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.4 | 6.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.4 | 5.6 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.4 | 3.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.4 | 2.0 | GO:0031437 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.4 | 2.8 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.4 | 1.2 | GO:1990117 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 0.4 | 2.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.4 | 2.3 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.4 | 5.0 | GO:0060068 | vagina development(GO:0060068) |
| 0.4 | 4.3 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.4 | 1.9 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.4 | 1.2 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.4 | 1.5 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.4 | 1.5 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.4 | 4.2 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.4 | 0.4 | GO:0071317 | cellular response to morphine(GO:0071315) cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.4 | 2.6 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.4 | 3.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.4 | 4.8 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.4 | 4.4 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) |
| 0.4 | 5.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.4 | 1.8 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.4 | 5.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.4 | 0.7 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.4 | 2.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.4 | 1.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.4 | 1.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.4 | 6.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.4 | 1.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.4 | 1.4 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.4 | 8.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.4 | 1.8 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.3 | 10.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.3 | 2.8 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.3 | 3.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 2.4 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.3 | 2.0 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.3 | 7.7 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.3 | 3.0 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.3 | 2.3 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.3 | 0.7 | GO:1905165 | regulation of lysosomal protein catabolic process(GO:1905165) |
| 0.3 | 1.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 2.6 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.3 | 1.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.3 | 1.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.3 | 3.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.3 | 32.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 5.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.3 | 2.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.3 | 2.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 | 3.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.3 | 1.8 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.3 | 12.0 | GO:0030815 | negative regulation of cAMP metabolic process(GO:0030815) |
| 0.3 | 11.9 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.3 | 0.9 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.3 | 3.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 0.9 | GO:1903626 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.3 | 1.2 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.3 | 0.9 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.3 | 1.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.3 | 0.9 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.3 | 3.1 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.3 | 0.8 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.3 | 1.9 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.3 | 9.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.3 | 11.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.3 | 11.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 1.3 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.3 | 0.8 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.3 | 1.0 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 1.5 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.3 | 1.8 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.3 | 1.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.3 | 2.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 3.8 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.3 | 0.5 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.3 | 0.5 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.3 | 1.8 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.3 | 4.0 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.3 | 1.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 1.5 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 3.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 0.7 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.2 | 6.0 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.4 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 2.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 0.5 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.2 | 1.2 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.2 | 6.6 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.2 | 3.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.2 | 1.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 3.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 7.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 0.7 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.2 | 3.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 0.9 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.2 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.2 | 2.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 0.8 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 6.6 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 1.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.2 | 1.8 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 5.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.2 | 0.8 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 2.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 1.8 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.2 | 0.6 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.2 | 4.9 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.2 | 2.3 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.2 | 3.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 5.1 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 3.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 3.4 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) observational learning(GO:0098597) |
| 0.2 | 0.8 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.2 | 0.9 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 0.6 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.2 | 1.7 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.2 | 1.3 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.2 | 2.0 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.2 | 0.4 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.2 | 1.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 3.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 1.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 1.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.2 | 4.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 6.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 3.1 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.2 | 3.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) chemical homeostasis within a tissue(GO:0048875) |
| 0.2 | 1.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 2.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.2 | 0.3 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.2 | 0.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.2 | 1.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.2 | 1.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 6.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 0.5 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.2 | 0.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 1.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 1.2 | GO:0098706 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 0.9 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.2 | 3.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 0.9 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 2.0 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.2 | 1.5 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.6 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 | 6.3 | GO:0090102 | cochlea development(GO:0090102) |
| 0.1 | 0.9 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 5.3 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.1 | 2.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.3 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.9 | GO:2000124 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.1 | 0.6 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 1.0 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 10.1 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 2.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 2.9 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 3.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.5 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 1.3 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.1 | 1.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.4 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.1 | 3.0 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.1 | 1.2 | GO:0030828 | positive regulation of cGMP metabolic process(GO:0030825) positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 7.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 2.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.7 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 1.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.8 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 0.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.7 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 1.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.7 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 4.7 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.1 | 0.3 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.1 | 1.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.2 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 0.8 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.8 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 6.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 1.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.6 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.6 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 1.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.5 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.1 | 0.3 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 2.0 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 0.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 6.2 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.1 | 0.3 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.1 | 0.7 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.4 | GO:2001107 | negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.1 | 0.8 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 1.8 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 7.2 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) |
| 0.1 | 0.3 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.1 | 0.7 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.5 | GO:0048866 | stem cell fate specification(GO:0048866) endocardium formation(GO:0060214) |
| 0.1 | 0.8 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 7.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 2.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.3 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.1 | 1.4 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 2.8 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 0.3 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.3 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.1 | 1.7 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.4 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 1.0 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.5 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.4 | GO:0036257 | multivesicular body organization(GO:0036257) multivesicular body assembly(GO:0036258) |
| 0.1 | 0.4 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.5 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 1.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 4.5 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.1 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.1 | 0.3 | GO:1900224 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.3 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.1 | 0.5 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.8 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.1 | 2.3 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 1.0 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 1.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.4 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 0.7 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 1.1 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.1 | 0.9 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.1 | 2.0 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.1 | 2.8 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.1 | 0.5 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 3.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 1.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.8 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 1.1 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 1.4 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 6.5 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 2.3 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.1 | 0.2 | GO:0070781 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.1 | 6.7 | GO:0050905 | neuromuscular process(GO:0050905) |
| 0.1 | 1.3 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 3.2 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.1 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 1.2 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 3.8 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.5 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 8.7 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.8 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 1.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 4.1 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.9 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 3.8 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 2.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.2 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.7 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 1.3 | GO:0007270 | neuron-neuron synaptic transmission(GO:0007270) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 3.7 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.4 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.6 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 1.1 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 1.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.3 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 1.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 3.3 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 1.4 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 0.3 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.5 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.6 | GO:0070849 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 1.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.5 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.0 | 0.1 | GO:1902188 | positive regulation of viral release from host cell(GO:1902188) |
| 0.0 | 0.1 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.2 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.3 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.6 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 1.8 | 5.3 | GO:0098855 | HCN channel complex(GO:0098855) |
| 1.3 | 9.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.3 | 19.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.2 | 3.5 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 1.2 | 3.5 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 1.1 | 4.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.0 | 6.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.0 | 5.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.0 | 7.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.0 | 4.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.9 | 10.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.9 | 0.9 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.9 | 13.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.9 | 16.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.8 | 3.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.8 | 4.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.8 | 3.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.7 | 7.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.7 | 7.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.7 | 2.2 | GO:0097635 | extrinsic component of pre-autophagosomal structure membrane(GO:0097632) extrinsic component of autophagosome membrane(GO:0097635) |
| 0.7 | 19.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.7 | 34.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.7 | 6.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.7 | 34.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.6 | 3.8 | GO:0097433 | dense body(GO:0097433) |
| 0.6 | 29.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.6 | 17.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.6 | 3.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.6 | 9.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 14.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.5 | 2.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.5 | 7.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.5 | 36.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.5 | 4.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 5.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.5 | 13.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.5 | 3.8 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.5 | 1.9 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.5 | 3.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.4 | 14.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.4 | 5.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 20.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.4 | 1.3 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.4 | 3.2 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.4 | 3.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.4 | 1.2 | GO:0097144 | BAX complex(GO:0097144) |
| 0.4 | 5.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 3.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.4 | 1.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.4 | 4.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 5.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 8.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 1.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 6.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 2.5 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.3 | 2.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 1.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 0.8 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 3.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 1.9 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.3 | 2.9 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.3 | 1.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.3 | 3.6 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.3 | 4.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 13.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 6.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 3.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 0.5 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.2 | 1.4 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 16.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 20.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 0.9 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.2 | 0.9 | GO:1990794 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) photoreceptor cell terminal bouton(GO:1990796) |
| 0.2 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 11.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 3.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 4.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 3.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 1.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 2.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 19.3 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.2 | 9.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 1.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 1.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.2 | 44.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 4.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 1.2 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.2 | 1.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 11.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.2 | 7.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 1.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 1.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 2.7 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.2 | 2.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 4.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 10.3 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.2 | 25.3 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 2.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 4.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 32.0 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 0.9 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 3.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.1 | 1.1 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 1.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 9.8 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 1.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 5.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.4 | GO:0071149 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 2.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 1.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 3.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 2.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 0.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 15.0 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 1.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 3.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 1.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.8 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 15.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 0.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.5 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 16.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 0.6 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 3.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 11.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 2.4 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 3.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 4.5 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 12.0 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 5.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 3.1 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 0.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 22.0 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 1.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 4.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 2.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 3.6 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 148.0 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.3 | GO:0098798 | mitochondrial protein complex(GO:0098798) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.7 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 3.7 | 11.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 3.5 | 10.5 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 3.1 | 12.6 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 2.5 | 7.6 | GO:0016662 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitric oxide binding(GO:0070026) |
| 2.4 | 7.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 2.1 | 12.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 2.0 | 12.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 2.0 | 19.8 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 1.9 | 11.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.9 | 9.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 1.6 | 4.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 1.6 | 9.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 1.5 | 7.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.5 | 6.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 1.5 | 6.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 1.5 | 11.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 1.4 | 4.3 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.4 | 4.3 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 1.4 | 9.5 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 1.3 | 12.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 1.3 | 4.0 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.3 | 5.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 1.2 | 3.7 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 1.2 | 7.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.2 | 8.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 1.1 | 19.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 1.1 | 4.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 1.0 | 11.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.0 | 3.0 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.9 | 3.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.9 | 4.5 | GO:2001070 | starch binding(GO:2001070) |
| 0.9 | 7.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.9 | 2.7 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.9 | 2.7 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.9 | 8.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.9 | 5.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.9 | 7.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.9 | 13.7 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.8 | 5.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.8 | 1.6 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.8 | 6.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.8 | 7.8 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.8 | 3.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.8 | 10.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.8 | 3.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.7 | 16.4 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.7 | 2.9 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.7 | 2.9 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.7 | 4.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.7 | 4.8 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.7 | 22.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.7 | 7.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.6 | 16.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.6 | 1.9 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.6 | 3.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.6 | 3.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.6 | 1.9 | GO:0005287 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.6 | 5.6 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.6 | 1.9 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.6 | 1.8 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.6 | 1.8 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.6 | 2.4 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.6 | 2.3 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.6 | 3.4 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.6 | 1.7 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.6 | 2.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.6 | 3.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.6 | 3.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.5 | 3.8 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.5 | 2.7 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.5 | 3.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.5 | 1.6 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.5 | 2.6 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.5 | 1.6 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.5 | 4.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.5 | 3.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.5 | 3.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.5 | 2.5 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.5 | 3.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.5 | 1.9 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.5 | 2.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.5 | 1.9 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.5 | 5.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.5 | 2.9 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.5 | 9.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.5 | 1.9 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.5 | 2.3 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.5 | 1.8 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.5 | 4.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.4 | 2.7 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.4 | 3.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.4 | 2.6 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.4 | 1.3 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.4 | 10.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.4 | 3.5 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.4 | 9.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.4 | 6.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.4 | 3.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 3.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.4 | 2.0 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.4 | 4.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 12.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.4 | 4.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.4 | 2.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.4 | 0.4 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.4 | 7.9 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.4 | 24.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 4.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.4 | 2.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.4 | 25.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.4 | 35.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.3 | 2.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.3 | 1.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.3 | 11.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.3 | 10.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 2.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 20.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.3 | 2.0 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.3 | 3.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 1.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.3 | 1.3 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.3 | 2.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.3 | 5.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 4.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 0.9 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.3 | 3.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 7.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 1.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.3 | 1.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 2.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 1.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 9.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 11.7 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.3 | 5.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 1.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.3 | 1.8 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 3.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.7 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 1.6 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 2.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 4.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 1.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 1.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 2.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 0.9 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.2 | 2.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 21.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 1.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 1.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 0.6 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 2.8 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 2.9 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 0.8 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.2 | 1.5 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.2 | 1.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 1.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 6.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 3.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 0.6 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.2 | 3.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 1.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 3.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 0.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 2.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 1.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 0.8 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.2 | 1.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 0.6 | GO:0047945 | L-phenylalanine:pyruvate aminotransferase activity(GO:0047312) glutamine-phenylpyruvate transaminase activity(GO:0047316) L-glutamine:pyruvate aminotransferase activity(GO:0047945) |
| 0.2 | 3.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 1.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 0.9 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 0.7 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.2 | 1.5 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.2 | 1.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 3.1 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.2 | 1.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.2 | 0.7 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.2 | 1.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 7.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 4.1 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.2 | 5.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 1.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 2.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 4.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 2.0 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 1.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 5.6 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 6.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 3.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 7.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 1.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 4.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 2.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.8 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 2.9 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 6.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 4.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 9.3 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 2.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 8.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.6 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.6 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 2.8 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.3 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 4.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 4.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.0 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 1.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 7.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 7.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 3.2 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.1 | 0.4 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.1 | 2.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.3 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 8.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 3.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.8 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 2.1 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 3.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 1.0 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 2.0 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.4 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.8 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.9 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.7 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 5.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.3 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 1.6 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.8 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 2.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.5 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 1.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 14.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 3.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 2.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 1.3 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 3.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 2.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.2 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.1 | 8.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 3.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 12.9 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.1 | 7.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 11.3 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.1 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.2 | GO:0004077 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.1 | 1.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 7.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 16.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.5 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 1.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 5.2 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 1.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.7 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 3.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 2.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.7 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 1.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0015093 | iron ion transmembrane transporter activity(GO:0005381) ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.2 | GO:1901567 | icosanoid binding(GO:0050542) icosatetraenoic acid binding(GO:0050543) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.3 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 4.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 1.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.4 | 4.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.4 | 2.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.3 | 6.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 21.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 8.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 6.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 10.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 19.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 2.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 13.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 11.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 5.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 16.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 2.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 3.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 3.8 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.2 | 5.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 7.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 0.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 4.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 3.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 4.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 23.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 1.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 3.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 13.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 2.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.9 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 20.9 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 1.5 | 9.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 1.0 | 24.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.8 | 9.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.7 | 15.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.7 | 10.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.6 | 8.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.5 | 16.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.5 | 4.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.4 | 15.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 8.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.4 | 13.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.4 | 5.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 4.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.3 | 4.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 48.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.3 | 10.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.3 | 5.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.3 | 4.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 10.7 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.3 | 12.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.3 | 13.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 6.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 4.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 3.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 4.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 6.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 3.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 5.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 3.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 3.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 3.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 10.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 5.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 4.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 7.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 8.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 4.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 3.1 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.2 | 3.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 3.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 4.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 5.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 6.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 3.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 1.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 3.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.8 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 3.0 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 3.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 4.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 20.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 3.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 15.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 15.3 | REACTOME NEURONAL SYSTEM | Genes involved in Neuronal System |
| 0.1 | 1.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 1.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.6 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 4.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.4 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 1.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 1.1 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.1 | 1.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 4.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 2.3 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 1.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 2.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 1.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.9 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 3.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |