Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
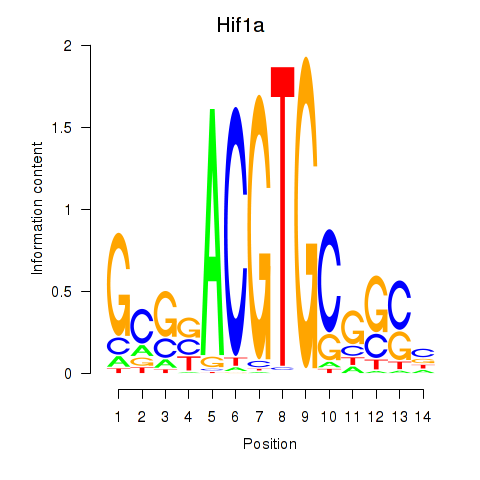
Results for Hif1a
Z-value: 1.40
Transcription factors associated with Hif1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hif1a
|
ENSMUSG00000021109.14 | Hif1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hif1a | mm39_v1_chr12_+_73954678_73954789 | -0.43 | 1.5e-04 | Click! |
Activity profile of Hif1a motif
Sorted Z-values of Hif1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hif1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_84938350 | 17.02 |
ENSMUST00000059383.8
ENSMUST00000216889.2 |
Fhl4
|
four and a half LIM domains 4 |
| chr4_-_55532453 | 16.03 |
ENSMUST00000132746.2
ENSMUST00000107619.3 |
Klf4
|
Kruppel-like factor 4 (gut) |
| chr18_+_64473091 | 13.74 |
ENSMUST00000175965.10
|
Onecut2
|
one cut domain, family member 2 |
| chr17_+_28988354 | 13.28 |
ENSMUST00000233109.2
ENSMUST00000004986.14 |
Mapk13
|
mitogen-activated protein kinase 13 |
| chr17_+_28988271 | 12.93 |
ENSMUST00000233984.2
ENSMUST00000233460.2 |
Mapk13
|
mitogen-activated protein kinase 13 |
| chr7_+_46495256 | 11.21 |
ENSMUST00000048209.16
ENSMUST00000210815.2 ENSMUST00000125862.8 ENSMUST00000210968.2 ENSMUST00000092621.12 ENSMUST00000210467.2 |
Ldha
|
lactate dehydrogenase A |
| chr7_+_28240262 | 10.86 |
ENSMUST00000119180.4
|
Sycn
|
syncollin |
| chr1_-_75156993 | 10.39 |
ENSMUST00000027396.15
|
Abcb6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr4_-_43523595 | 9.98 |
ENSMUST00000107914.10
|
Tpm2
|
tropomyosin 2, beta |
| chr17_-_43941157 | 9.52 |
ENSMUST00000045717.7
ENSMUST00000233077.2 ENSMUST00000168073.9 |
Tdrd6
|
tudor domain containing 6 |
| chr15_+_80556023 | 9.38 |
ENSMUST00000023044.7
|
Fam83f
|
family with sequence similarity 83, member F |
| chr1_+_172309337 | 9.30 |
ENSMUST00000127052.8
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr7_+_142660971 | 8.66 |
ENSMUST00000009689.11
|
Kcnq1
|
potassium voltage-gated channel, subfamily Q, member 1 |
| chr1_+_172309766 | 8.40 |
ENSMUST00000135267.8
ENSMUST00000052629.13 ENSMUST00000111235.8 |
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr2_+_167922924 | 7.77 |
ENSMUST00000052125.7
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr10_+_43355113 | 7.54 |
ENSMUST00000040147.8
|
Bend3
|
BEN domain containing 3 |
| chr7_+_46495521 | 7.37 |
ENSMUST00000133062.2
|
Ldha
|
lactate dehydrogenase A |
| chr2_-_14060774 | 7.17 |
ENSMUST00000114753.8
ENSMUST00000091429.12 |
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr2_-_14060840 | 7.08 |
ENSMUST00000074854.9
|
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr11_-_88608958 | 7.02 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr10_-_80223475 | 6.81 |
ENSMUST00000105350.3
|
Mex3d
|
mex3 RNA binding family member D |
| chr12_+_110413523 | 6.54 |
ENSMUST00000222276.2
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr11_-_88609048 | 6.45 |
ENSMUST00000107909.8
|
Msi2
|
musashi RNA-binding protein 2 |
| chr3_-_131196213 | 6.14 |
ENSMUST00000197057.2
|
Sgms2
|
sphingomyelin synthase 2 |
| chr11_+_61575245 | 6.09 |
ENSMUST00000093019.6
|
Fam83g
|
family with sequence similarity 83, member G |
| chr17_+_46991972 | 5.95 |
ENSMUST00000002845.8
|
Mea1
|
male enhanced antigen 1 |
| chr14_-_20231871 | 5.64 |
ENSMUST00000024011.10
|
Kcnk5
|
potassium channel, subfamily K, member 5 |
| chr17_+_27276262 | 5.60 |
ENSMUST00000049308.9
|
Itpr3
|
inositol 1,4,5-triphosphate receptor 3 |
| chr19_+_6952580 | 5.45 |
ENSMUST00000237084.2
ENSMUST00000236218.2 ENSMUST00000237235.2 |
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr10_+_43354807 | 5.41 |
ENSMUST00000167488.9
|
Bend3
|
BEN domain containing 3 |
| chr5_+_129097133 | 5.15 |
ENSMUST00000031383.14
ENSMUST00000111343.2 |
Ran
|
RAN, member RAS oncogene family |
| chr7_-_98831916 | 5.14 |
ENSMUST00000033001.6
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr11_-_72302520 | 5.09 |
ENSMUST00000108500.8
ENSMUST00000050226.7 |
Smtnl2
|
smoothelin-like 2 |
| chr9_-_95727267 | 5.06 |
ENSMUST00000093800.9
|
Pls1
|
plastin 1 (I-isoform) |
| chr17_+_46991926 | 5.04 |
ENSMUST00000233491.2
|
Mea1
|
male enhanced antigen 1 |
| chr2_+_153334710 | 5.02 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr17_+_46991906 | 4.95 |
ENSMUST00000233430.2
|
Mea1
|
male enhanced antigen 1 |
| chr1_+_172327812 | 4.93 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr11_+_120839879 | 4.86 |
ENSMUST00000154187.8
ENSMUST00000100130.10 ENSMUST00000129473.8 ENSMUST00000168579.8 |
Slc16a3
|
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
| chr9_-_58220469 | 4.86 |
ENSMUST00000061799.10
|
Loxl1
|
lysyl oxidase-like 1 |
| chr9_+_107464841 | 4.81 |
ENSMUST00000010192.11
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr19_+_6952319 | 4.73 |
ENSMUST00000070850.8
|
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr15_-_31453708 | 4.72 |
ENSMUST00000110408.3
|
Ropn1l
|
ropporin 1-like |
| chr1_+_55127110 | 4.64 |
ENSMUST00000075242.7
|
Hspe1
|
heat shock protein 1 (chaperonin 10) |
| chr8_+_91635192 | 4.63 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr15_-_36609208 | 4.51 |
ENSMUST00000001809.15
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr16_+_70111007 | 4.47 |
ENSMUST00000170464.3
|
Gbe1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr9_-_35028100 | 4.44 |
ENSMUST00000034537.8
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr10_+_75729237 | 4.32 |
ENSMUST00000009236.6
ENSMUST00000217811.2 |
Derl3
|
Der1-like domain family, member 3 |
| chr11_-_102709936 | 4.30 |
ENSMUST00000068933.12
|
Gjc1
|
gap junction protein, gamma 1 |
| chr6_-_38614155 | 4.29 |
ENSMUST00000096030.7
ENSMUST00000201345.2 |
Klrg2
|
killer cell lectin-like receptor subfamily G, member 2 |
| chr17_+_46992101 | 4.20 |
ENSMUST00000233612.2
|
Mea1
|
male enhanced antigen 1 |
| chr16_+_93885775 | 4.20 |
ENSMUST00000072182.9
|
Sim2
|
single-minded family bHLH transcription factor 2 |
| chr19_+_4560500 | 4.07 |
ENSMUST00000068004.13
ENSMUST00000224726.3 |
Pcx
|
pyruvate carboxylase |
| chr19_-_44058175 | 3.94 |
ENSMUST00000172041.8
ENSMUST00000071698.13 ENSMUST00000112028.10 |
Erlin1
|
ER lipid raft associated 1 |
| chr16_+_70110837 | 3.92 |
ENSMUST00000163832.8
|
Gbe1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr15_-_75781387 | 3.88 |
ENSMUST00000123712.8
ENSMUST00000141475.2 ENSMUST00000144614.8 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr16_+_70110975 | 3.88 |
ENSMUST00000023393.15
|
Gbe1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr17_+_46991350 | 3.79 |
ENSMUST00000232732.2
|
Mea1
|
male enhanced antigen 1 |
| chr3_-_101017594 | 3.78 |
ENSMUST00000102694.4
|
Ptgfrn
|
prostaglandin F2 receptor negative regulator |
| chr11_-_116001037 | 3.70 |
ENSMUST00000106441.8
ENSMUST00000021120.6 |
Trim47
|
tripartite motif-containing 47 |
| chr6_-_70769135 | 3.66 |
ENSMUST00000066134.6
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr15_-_102112159 | 3.64 |
ENSMUST00000229252.2
ENSMUST00000229770.2 |
Csad
|
cysteine sulfinic acid decarboxylase |
| chr8_+_84724130 | 3.62 |
ENSMUST00000095228.5
|
Samd1
|
sterile alpha motif domain containing 1 |
| chr19_-_31742427 | 3.61 |
ENSMUST00000065067.14
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr11_+_72851989 | 3.55 |
ENSMUST00000163326.8
ENSMUST00000108485.9 ENSMUST00000021142.8 ENSMUST00000108486.8 ENSMUST00000108484.8 |
Atp2a3
|
ATPase, Ca++ transporting, ubiquitous |
| chr2_-_163261439 | 3.48 |
ENSMUST00000046908.10
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr5_+_108842294 | 3.46 |
ENSMUST00000013633.12
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr3_+_122039274 | 3.35 |
ENSMUST00000178826.8
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr4_-_21685781 | 3.32 |
ENSMUST00000076206.11
|
Prdm13
|
PR domain containing 13 |
| chr5_+_108842088 | 3.31 |
ENSMUST00000197255.2
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr1_-_55127183 | 3.30 |
ENSMUST00000027123.15
|
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr12_+_24701273 | 3.30 |
ENSMUST00000020982.7
|
Klf11
|
Kruppel-like factor 11 |
| chr6_-_100264439 | 3.27 |
ENSMUST00000101118.4
|
Rybp
|
RING1 and YY1 binding protein |
| chr15_-_75781138 | 3.23 |
ENSMUST00000145764.2
ENSMUST00000116440.9 ENSMUST00000151066.8 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr6_+_108805594 | 3.22 |
ENSMUST00000089162.5
|
Edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr15_-_36609812 | 3.21 |
ENSMUST00000226496.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr3_-_75864195 | 3.19 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr19_-_44057800 | 3.14 |
ENSMUST00000170801.8
|
Erlin1
|
ER lipid raft associated 1 |
| chr1_+_74811045 | 3.09 |
ENSMUST00000006716.8
|
Wnt6
|
wingless-type MMTV integration site family, member 6 |
| chr15_-_75781168 | 3.09 |
ENSMUST00000089680.10
ENSMUST00000141268.8 ENSMUST00000023235.13 ENSMUST00000109972.9 ENSMUST00000089681.12 ENSMUST00000109975.10 ENSMUST00000154584.9 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr9_+_107446310 | 3.05 |
ENSMUST00000010191.13
ENSMUST00000193747.2 |
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr6_+_88701810 | 3.00 |
ENSMUST00000089449.5
|
Mgll
|
monoglyceride lipase |
| chr5_+_108842125 | 2.93 |
ENSMUST00000112560.8
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr5_+_77414031 | 2.88 |
ENSMUST00000113449.2
|
Rest
|
RE1-silencing transcription factor |
| chr10_+_95776543 | 2.86 |
ENSMUST00000053484.8
|
Eea1
|
early endosome antigen 1 |
| chr7_-_68398989 | 2.84 |
ENSMUST00000048068.15
|
Arrdc4
|
arrestin domain containing 4 |
| chr1_-_36722195 | 2.82 |
ENSMUST00000170295.8
ENSMUST00000114981.3 |
Fam178b
|
family with sequence similarity 178, member B |
| chr7_-_68398917 | 2.81 |
ENSMUST00000118110.3
|
Arrdc4
|
arrestin domain containing 4 |
| chr12_+_32870334 | 2.80 |
ENSMUST00000020886.9
|
Nampt
|
nicotinamide phosphoribosyltransferase |
| chr10_+_17598961 | 2.79 |
ENSMUST00000038107.9
ENSMUST00000219558.2 ENSMUST00000218370.2 |
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chrX_+_10583629 | 2.66 |
ENSMUST00000115524.8
ENSMUST00000008179.7 ENSMUST00000156321.2 |
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr1_+_78286946 | 2.54 |
ENSMUST00000036172.10
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr19_+_25587896 | 2.54 |
ENSMUST00000048935.6
|
Dmrt3
|
doublesex and mab-3 related transcription factor 3 |
| chr3_+_122039206 | 2.54 |
ENSMUST00000029769.14
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr8_-_123278054 | 2.53 |
ENSMUST00000156333.9
ENSMUST00000067252.14 |
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr2_+_68691778 | 2.39 |
ENSMUST00000028426.9
|
Cers6
|
ceramide synthase 6 |
| chr8_+_34006758 | 2.32 |
ENSMUST00000149399.8
|
Tex15
|
testis expressed gene 15 |
| chr16_+_70110944 | 2.29 |
ENSMUST00000171132.8
|
Gbe1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr19_-_44058129 | 2.26 |
ENSMUST00000169092.2
|
Erlin1
|
ER lipid raft associated 1 |
| chr14_-_21102487 | 2.24 |
ENSMUST00000154460.8
ENSMUST00000130291.8 |
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr6_-_86503178 | 2.21 |
ENSMUST00000053015.7
|
Pcbp1
|
poly(rC) binding protein 1 |
| chr4_-_129494435 | 2.20 |
ENSMUST00000102593.11
|
Eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr8_+_34222058 | 2.18 |
ENSMUST00000167264.8
ENSMUST00000187392.7 |
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr11_+_60428788 | 2.12 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr4_+_152410291 | 2.12 |
ENSMUST00000103191.11
ENSMUST00000139685.8 ENSMUST00000188151.2 |
Rpl22
|
ribosomal protein L22 |
| chr8_+_34221861 | 2.07 |
ENSMUST00000170705.8
|
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr9_+_108225026 | 2.07 |
ENSMUST00000035237.12
ENSMUST00000194959.6 |
Usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr8_+_34222266 | 1.98 |
ENSMUST00000190675.2
ENSMUST00000171010.8 |
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr2_+_74593324 | 1.98 |
ENSMUST00000047793.6
|
Hoxd1
|
homeobox D1 |
| chr1_+_120530134 | 1.97 |
ENSMUST00000079721.9
|
En1
|
engrailed 1 |
| chr4_-_62438122 | 1.97 |
ENSMUST00000107444.8
ENSMUST00000030090.4 |
Alad
|
aminolevulinate, delta-, dehydratase |
| chr7_+_98138904 | 1.92 |
ENSMUST00000205956.2
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr10_+_80100812 | 1.84 |
ENSMUST00000105362.8
ENSMUST00000105361.10 |
Dazap1
|
DAZ associated protein 1 |
| chr13_-_53135064 | 1.82 |
ENSMUST00000071065.8
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr12_+_104998895 | 1.80 |
ENSMUST00000223244.2
ENSMUST00000021522.5 |
Glrx5
|
glutaredoxin 5 |
| chr5_-_148988413 | 1.78 |
ENSMUST00000093196.11
|
Hmgb1
|
high mobility group box 1 |
| chr10_+_84938452 | 1.78 |
ENSMUST00000095383.6
|
Tmem263
|
transmembrane protein 263 |
| chr2_-_27317004 | 1.74 |
ENSMUST00000056176.8
|
Vav2
|
vav 2 oncogene |
| chr4_-_43045685 | 1.73 |
ENSMUST00000107956.8
ENSMUST00000107957.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr14_+_21102642 | 1.73 |
ENSMUST00000045376.11
|
Adk
|
adenosine kinase |
| chr4_-_118148471 | 1.61 |
ENSMUST00000222620.2
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F |
| chr14_-_30740946 | 1.61 |
ENSMUST00000228341.2
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr17_-_46991709 | 1.61 |
ENSMUST00000233524.2
ENSMUST00000233733.2 ENSMUST00000071841.7 ENSMUST00000165007.9 |
Klhdc3
|
kelch domain containing 3 |
| chr5_-_144902598 | 1.58 |
ENSMUST00000110677.8
ENSMUST00000085684.11 ENSMUST00000100461.7 |
Smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr7_-_136915602 | 1.53 |
ENSMUST00000210774.2
|
Ebf3
|
early B cell factor 3 |
| chr14_-_30741012 | 1.52 |
ENSMUST00000037739.8
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr4_-_43046196 | 1.52 |
ENSMUST00000036462.12
|
Fam214b
|
family with sequence similarity 214, member B |
| chr4_+_116734573 | 1.52 |
ENSMUST00000044823.4
|
Zswim5
|
zinc finger SWIM-type containing 5 |
| chr5_+_135197228 | 1.51 |
ENSMUST00000111187.10
ENSMUST00000111188.5 ENSMUST00000202606.3 |
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr14_+_21102662 | 1.45 |
ENSMUST00000223915.2
|
Adk
|
adenosine kinase |
| chr17_-_23896394 | 1.45 |
ENSMUST00000233428.2
ENSMUST00000233814.2 ENSMUST00000167059.9 ENSMUST00000024698.10 |
Tnfrsf12a
|
tumor necrosis factor receptor superfamily, member 12a |
| chr15_+_85942928 | 1.41 |
ENSMUST00000138134.8
|
Gramd4
|
GRAM domain containing 4 |
| chr8_+_105318067 | 1.40 |
ENSMUST00000059588.8
|
Pdp2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr4_-_129494378 | 1.40 |
ENSMUST00000135055.8
|
Eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr17_-_26314438 | 1.39 |
ENSMUST00000236547.2
|
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr17_-_26314461 | 1.37 |
ENSMUST00000236128.2
ENSMUST00000025007.7 |
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr2_+_155359868 | 1.35 |
ENSMUST00000029135.15
ENSMUST00000065973.9 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr13_+_81805941 | 1.33 |
ENSMUST00000049055.8
|
Lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr7_+_98138490 | 1.32 |
ENSMUST00000205937.2
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr6_+_88701394 | 1.32 |
ENSMUST00000113585.9
|
Mgll
|
monoglyceride lipase |
| chr19_-_44994824 | 1.26 |
ENSMUST00000097715.4
|
Mrpl43
|
mitochondrial ribosomal protein L43 |
| chr2_+_27405169 | 1.25 |
ENSMUST00000113952.10
|
Wdr5
|
WD repeat domain 5 |
| chr11_-_120238917 | 1.25 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr14_-_31552335 | 1.24 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr13_+_81805782 | 1.23 |
ENSMUST00000224300.2
ENSMUST00000224592.2 |
Lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr6_-_83433357 | 1.23 |
ENSMUST00000186548.7
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr13_-_30168374 | 1.20 |
ENSMUST00000221536.2
ENSMUST00000222730.2 |
E2f3
|
E2F transcription factor 3 |
| chr16_-_10994135 | 1.20 |
ENSMUST00000037633.16
|
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr10_+_80100868 | 1.18 |
ENSMUST00000092305.6
|
Dazap1
|
DAZ associated protein 1 |
| chr2_+_127967951 | 1.16 |
ENSMUST00000089634.12
ENSMUST00000019281.14 ENSMUST00000110341.9 ENSMUST00000103211.8 ENSMUST00000103210.2 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chr11_-_95956176 | 1.13 |
ENSMUST00000100528.5
|
Ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr3_-_58599812 | 1.12 |
ENSMUST00000070368.8
|
Siah2
|
siah E3 ubiquitin protein ligase 2 |
| chr11_+_102080446 | 1.11 |
ENSMUST00000070334.10
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr15_-_57755753 | 1.11 |
ENSMUST00000022993.7
|
Derl1
|
Der1-like domain family, member 1 |
| chr1_-_180083859 | 1.10 |
ENSMUST00000111108.10
|
Psen2
|
presenilin 2 |
| chr12_-_40087393 | 1.10 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr15_+_80862074 | 1.09 |
ENSMUST00000229727.2
|
Sgsm3
|
small G protein signaling modulator 3 |
| chr8_-_70426910 | 1.08 |
ENSMUST00000116463.4
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr6_+_88701578 | 1.07 |
ENSMUST00000150180.4
ENSMUST00000163271.8 |
Mgll
|
monoglyceride lipase |
| chr11_-_102710490 | 1.07 |
ENSMUST00000092567.11
|
Gjc1
|
gap junction protein, gamma 1 |
| chr9_-_110483210 | 1.05 |
ENSMUST00000196488.5
ENSMUST00000133191.8 ENSMUST00000167320.8 |
Nbeal2
|
neurobeachin-like 2 |
| chr13_-_112788829 | 1.04 |
ENSMUST00000075748.7
|
Ddx4
|
DEAD box helicase 4 |
| chr2_+_179761093 | 1.03 |
ENSMUST00000040668.9
|
Osbpl2
|
oxysterol binding protein-like 2 |
| chr9_-_57168777 | 1.02 |
ENSMUST00000217657.2
|
1700017B05Rik
|
RIKEN cDNA 1700017B05 gene |
| chr11_-_120239301 | 1.02 |
ENSMUST00000062147.14
ENSMUST00000128055.2 |
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr14_+_16182428 | 1.02 |
ENSMUST00000170104.3
|
Gm3411
|
predicted gene 3411 |
| chr5_-_123662175 | 1.00 |
ENSMUST00000200247.5
ENSMUST00000111586.8 ENSMUST00000031385.7 ENSMUST00000145152.8 ENSMUST00000111587.10 |
Diablo
|
diablo, IAP-binding mitochondrial protein |
| chr5_+_33815466 | 0.98 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr7_-_121666486 | 0.95 |
ENSMUST00000033159.4
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr11_-_120239339 | 0.93 |
ENSMUST00000071555.13
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr8_+_111821262 | 0.90 |
ENSMUST00000135302.8
ENSMUST00000039333.10 |
Pdpr
|
pyruvate dehydrogenase phosphatase regulatory subunit |
| chr11_-_72380730 | 0.88 |
ENSMUST00000045303.10
|
Spns2
|
spinster homolog 2 |
| chr6_+_88701470 | 0.87 |
ENSMUST00000113581.8
|
Mgll
|
monoglyceride lipase |
| chr14_+_121148625 | 0.87 |
ENSMUST00000032898.9
|
Ipo5
|
importin 5 |
| chr1_-_161078723 | 0.86 |
ENSMUST00000051925.5
ENSMUST00000071718.12 |
Prdx6
|
peroxiredoxin 6 |
| chr2_+_162773440 | 0.83 |
ENSMUST00000130411.7
ENSMUST00000126163.3 |
Srsf6
|
serine and arginine-rich splicing factor 6 |
| chr5_+_45650821 | 0.82 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr13_-_72111832 | 0.77 |
ENSMUST00000077337.9
|
Irx1
|
Iroquois homeobox 1 |
| chr19_+_44994905 | 0.77 |
ENSMUST00000026227.3
|
Twnk
|
twinkle mtDNA helicase |
| chr5_-_135280063 | 0.76 |
ENSMUST00000062572.3
|
Fzd9
|
frizzled class receptor 9 |
| chr4_-_34882917 | 0.74 |
ENSMUST00000098163.9
ENSMUST00000047950.6 |
Zfp292
|
zinc finger protein 292 |
| chr4_+_129494463 | 0.73 |
ENSMUST00000102591.10
ENSMUST00000181579.8 ENSMUST00000173758.8 |
Tmem234
|
transmembrane protein 234 |
| chr11_-_88608920 | 0.72 |
ENSMUST00000092794.12
|
Msi2
|
musashi RNA-binding protein 2 |
| chr7_+_99808452 | 0.70 |
ENSMUST00000032967.4
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr4_+_101276893 | 0.70 |
ENSMUST00000131397.2
ENSMUST00000133055.8 |
Ak4
|
adenylate kinase 4 |
| chr5_+_135093056 | 0.68 |
ENSMUST00000071263.7
|
Dnajc30
|
DnaJ heat shock protein family (Hsp40) member C30 |
| chr4_-_57300361 | 0.67 |
ENSMUST00000153926.8
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr2_+_68691902 | 0.66 |
ENSMUST00000176018.2
|
Cers6
|
ceramide synthase 6 |
| chr8_-_46605196 | 0.64 |
ENSMUST00000110378.9
|
Snx25
|
sorting nexin 25 |
| chr13_-_112788890 | 0.64 |
ENSMUST00000099166.10
|
Ddx4
|
DEAD box helicase 4 |
| chr2_+_155360015 | 0.62 |
ENSMUST00000103142.12
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr3_+_152101698 | 0.62 |
ENSMUST00000200570.5
|
Zzz3
|
zinc finger, ZZ domain containing 3 |
| chr12_+_110413677 | 0.61 |
ENSMUST00000220509.2
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr16_+_32698470 | 0.61 |
ENSMUST00000232272.2
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr18_-_3337679 | 0.60 |
ENSMUST00000150235.8
ENSMUST00000154470.8 |
Crem
|
cAMP responsive element modulator |
| chr9_+_70450130 | 0.58 |
ENSMUST00000049263.9
|
Sltm
|
SAFB-like, transcription modulator |
| chr7_+_3706992 | 0.57 |
ENSMUST00000006496.15
ENSMUST00000108623.8 ENSMUST00000139818.2 ENSMUST00000108625.8 |
Rps9
|
ribosomal protein S9 |
| chr11_+_117006020 | 0.57 |
ENSMUST00000103026.10
ENSMUST00000090433.6 |
Sec14l1
|
SEC14-like lipid binding 1 |
| chr10_-_13068900 | 0.57 |
ENSMUST00000019950.6
|
Ltv1
|
LTV1 ribosome biogenesis factor |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 16.0 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 2.9 | 8.7 | GO:0072347 | response to anesthetic(GO:0072347) |
| 1.9 | 18.6 | GO:0019660 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 1.7 | 5.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 1.6 | 13.0 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 1.5 | 10.4 | GO:0015886 | heme transport(GO:0015886) |
| 1.3 | 5.4 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 1.3 | 5.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.1 | 7.7 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 1.1 | 3.2 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.1 | 3.2 | GO:0006175 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 1.0 | 5.8 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.9 | 5.6 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.9 | 3.7 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.9 | 2.6 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.8 | 9.3 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.8 | 4.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.8 | 3.0 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.7 | 3.6 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.7 | 3.6 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.7 | 2.9 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.7 | 2.8 | GO:0035801 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.7 | 13.7 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.7 | 2.0 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.7 | 5.9 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.6 | 6.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.6 | 1.8 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.6 | 5.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.6 | 14.3 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.5 | 1.6 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.5 | 2.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.4 | 9.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.4 | 0.4 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.4 | 1.2 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.4 | 4.9 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.4 | 4.3 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.4 | 2.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.3 | 3.1 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) positive regulation of tooth mineralization(GO:0070172) |
| 0.3 | 1.8 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.3 | 1.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.3 | 5.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 2.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.3 | 1.6 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.3 | 0.8 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.2 | 1.2 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.2 | 1.2 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.2 | 4.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 | 3.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.2 | 10.2 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.2 | 24.5 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.2 | 1.1 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.2 | 2.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.2 | 2.8 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.2 | 1.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 14.6 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 3.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 8.3 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.2 | 0.5 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.2 | 2.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 5.1 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.2 | 1.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 0.8 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 0.2 | 3.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 1.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 2.5 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 | 1.0 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 2.5 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 4.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 1.3 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.9 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 2.7 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 0.5 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 17.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.0 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 4.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 3.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 2.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.4 | GO:0009223 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 1.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 2.5 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.1 | 0.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.6 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 4.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.5 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 1.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 3.0 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.8 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 1.7 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 4.2 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.1 | 6.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 7.8 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.1 | 3.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 1.0 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.1 | 0.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 13.0 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 0.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.5 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 28.8 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 2.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 3.2 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 5.3 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 4.0 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.5 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 2.5 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 8.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.3 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 1.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 2.0 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 3.6 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 1.1 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.6 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 2.0 | 5.9 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 1.7 | 5.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 1.5 | 10.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.8 | 5.4 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.7 | 10.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.6 | 18.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.6 | 2.9 | GO:0044308 | axonal spine(GO:0044308) |
| 0.6 | 5.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.6 | 11.2 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.4 | 17.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 3.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.3 | 2.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 7.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.3 | 16.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.3 | 7.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 9.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 5.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 16.7 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 7.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 5.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 5.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 13.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 27.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 10.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 3.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 3.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 1.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 3.2 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.6 | GO:0034448 | EGO complex(GO:0034448) |
| 0.1 | 1.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 2.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 9.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 1.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 8.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.2 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 1.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 7.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 9.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.8 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 4.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 9.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 4.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 4.4 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.3 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 3.6 | 14.6 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 3.2 | 9.7 | GO:0001571 | non-tyrosine kinase fibroblast growth factor receptor activity(GO:0001571) |
| 3.2 | 16.0 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 2.0 | 5.9 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 1.7 | 10.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.7 | 18.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.4 | 5.6 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 1.3 | 5.4 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 1.3 | 5.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 1.2 | 8.7 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 1.2 | 13.0 | GO:0000182 | rDNA binding(GO:0000182) |
| 1.1 | 3.2 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 1.0 | 6.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.0 | 4.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.0 | 24.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.9 | 2.8 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.8 | 3.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.7 | 21.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.6 | 5.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.6 | 2.5 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.6 | 4.4 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.6 | 3.6 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.6 | 1.8 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.6 | 2.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.5 | 4.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.5 | 3.6 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.4 | 17.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.4 | 2.6 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.4 | 10.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 1.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 2.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.4 | 3.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.4 | 5.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 2.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.3 | 1.3 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.3 | 3.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 2.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 2.8 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 5.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 3.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 0.7 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 2.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 4.9 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.2 | 10.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 2.8 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 3.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 6.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.9 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 10.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 1.5 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 3.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 10.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 2.2 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 1.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 7.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.5 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 2.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.9 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 1.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.8 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 2.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 4.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 3.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.6 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 1.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.6 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.5 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 4.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 2.0 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 3.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 4.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 3.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 4.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 9.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 3.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 10.1 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 26.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 5.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 5.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 20.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 14.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 8.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 3.6 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 3.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 3.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 5.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 24.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 1.0 | 24.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.7 | 16.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.6 | 5.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 7.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.3 | 3.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 8.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 11.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.3 | 3.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 8.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 5.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 3.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 5.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 10.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 5.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.2 | 7.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 6.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 2.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 8.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 10.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 3.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 17.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 4.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.9 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 2.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 5.1 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 1.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 2.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 2.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 3.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 4.9 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 2.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |