Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Hinfp
Z-value: 0.88
Transcription factors associated with Hinfp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hinfp
|
ENSMUSG00000032119.6 | Hinfp |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hinfp | mm39_v1_chr9_-_44216892_44216986 | 0.26 | 2.9e-02 | Click! |
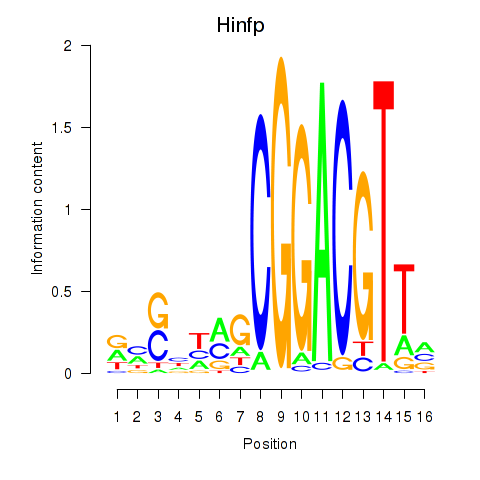
Activity profile of Hinfp motif
Sorted Z-values of Hinfp motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hinfp
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_92523396 | 4.24 |
ENSMUST00000209074.2
ENSMUST00000208356.2 ENSMUST00000032877.11 |
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr1_-_88133472 | 4.21 |
ENSMUST00000119972.4
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3 |
| chr1_-_128520002 | 4.11 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr17_-_14279793 | 3.96 |
ENSMUST00000186636.3
|
Gm7358
|
predicted gene 7358 |
| chr6_-_40906665 | 3.95 |
ENSMUST00000136499.2
|
1700074P13Rik
|
RIKEN cDNA 1700074P13 gene |
| chrX_+_156482116 | 3.58 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr2_-_156848923 | 3.50 |
ENSMUST00000146413.8
ENSMUST00000103129.9 ENSMUST00000103130.8 |
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr1_+_134890288 | 3.48 |
ENSMUST00000027687.8
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chrX_+_9713167 | 3.42 |
ENSMUST00000044987.3
|
H2ap
|
H2A.P histone |
| chr11_-_73029070 | 3.37 |
ENSMUST00000052140.3
|
Haspin
|
histone H3 associated protein kinase |
| chr7_-_45546081 | 3.28 |
ENSMUST00000120299.4
ENSMUST00000039049.16 |
Syngr4
|
synaptogyrin 4 |
| chr1_+_191553556 | 3.07 |
ENSMUST00000027931.8
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2 |
| chr6_+_29402868 | 3.05 |
ENSMUST00000154619.5
|
Ccdc136
|
coiled-coil domain containing 136 |
| chr3_+_68912043 | 2.93 |
ENSMUST00000042901.15
|
Smc4
|
structural maintenance of chromosomes 4 |
| chr7_-_126574959 | 2.90 |
ENSMUST00000206296.2
ENSMUST00000205437.3 |
Cdiptos
|
CDIP transferase, opposite strand |
| chr6_+_41090484 | 2.82 |
ENSMUST00000103267.3
|
Trbv12-1
|
T cell receptor beta, variable 12-1 |
| chrX_+_156481906 | 2.81 |
ENSMUST00000136141.2
ENSMUST00000190091.7 |
Smpx
|
small muscle protein, X-linked |
| chr7_+_121758646 | 2.80 |
ENSMUST00000033154.8
ENSMUST00000205901.2 |
Plk1
|
polo like kinase 1 |
| chr8_-_43594523 | 2.71 |
ENSMUST00000059692.4
|
Triml1
|
tripartite motif family-like 1 |
| chr11_-_53871121 | 2.71 |
ENSMUST00000076493.11
|
Slc22a21
|
solute carrier family 22 (organic cation transporter), member 21 |
| chr6_+_41520150 | 2.67 |
ENSMUST00000103295.2
|
Trbj2-3
|
T cell receptor beta joining 2-3 |
| chr2_+_29014984 | 2.63 |
ENSMUST00000061578.9
|
Setx
|
senataxin |
| chrX_+_85235370 | 2.60 |
ENSMUST00000026036.5
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr5_-_8472582 | 2.59 |
ENSMUST00000168500.8
ENSMUST00000002368.16 |
Dbf4
|
DBF4 zinc finger |
| chr6_+_41520737 | 2.54 |
ENSMUST00000103298.2
|
Trbj2-7
|
T cell receptor beta joining 2-7 |
| chr5_+_36641922 | 2.48 |
ENSMUST00000060100.3
|
Ccdc96
|
coiled-coil domain containing 96 |
| chr6_+_41520287 | 2.33 |
ENSMUST00000103296.2
|
Trbj2-4
|
T cell receptor beta joining 2-4 |
| chr1_-_171854818 | 2.31 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr5_-_8472696 | 2.29 |
ENSMUST00000171808.8
|
Dbf4
|
DBF4 zinc finger |
| chr5_+_137777111 | 2.28 |
ENSMUST00000126126.8
ENSMUST00000031739.6 |
Ppp1r35
|
protein phosphatase 1, regulatory subunit 35 |
| chr1_+_171216480 | 2.27 |
ENSMUST00000056449.9
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr11_+_3280771 | 2.26 |
ENSMUST00000136536.8
ENSMUST00000093399.11 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr13_-_21652632 | 2.25 |
ENSMUST00000068235.6
|
Nkapl
|
NFKB activating protein-like |
| chr15_-_31453708 | 2.23 |
ENSMUST00000110408.3
|
Ropn1l
|
ropporin 1-like |
| chr12_-_32111214 | 2.23 |
ENSMUST00000003079.12
ENSMUST00000036497.16 |
Prkar2b
|
protein kinase, cAMP dependent regulatory, type II beta |
| chrX_+_70599524 | 2.22 |
ENSMUST00000072699.13
ENSMUST00000114582.9 ENSMUST00000015361.11 ENSMUST00000088874.10 |
Hmgb3
|
high mobility group box 3 |
| chr8_-_57940834 | 2.21 |
ENSMUST00000034022.4
|
Sap30
|
sin3 associated polypeptide |
| chr17_+_33651864 | 2.20 |
ENSMUST00000174088.3
|
Actl9
|
actin-like 9 |
| chr5_+_25386452 | 2.14 |
ENSMUST00000030778.9
|
Galntl5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr5_-_113957362 | 2.13 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr4_-_133695264 | 2.04 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr5_+_33815910 | 2.04 |
ENSMUST00000114426.10
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr5_+_25386487 | 2.01 |
ENSMUST00000114965.2
|
Galntl5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr9_+_57847387 | 2.01 |
ENSMUST00000043059.9
|
Sema7a
|
sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A |
| chr5_+_124068728 | 2.00 |
ENSMUST00000094320.10
ENSMUST00000165148.4 |
Ccdc62
|
coiled-coil domain containing 62 |
| chr6_+_47930324 | 2.00 |
ENSMUST00000101445.11
|
Zfp956
|
zinc finger protein 956 |
| chr13_+_51799268 | 1.97 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr3_+_108847375 | 1.96 |
ENSMUST00000197427.5
ENSMUST00000059946.11 ENSMUST00000098680.9 |
Henmt1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr6_+_41520379 | 1.94 |
ENSMUST00000103297.2
|
Trbj2-5
|
T cell receptor beta joining 2-5 |
| chrY_+_1010543 | 1.93 |
ENSMUST00000091197.4
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr1_+_139382485 | 1.91 |
ENSMUST00000200083.5
ENSMUST00000053364.12 |
Aspm
|
abnormal spindle microtubule assembly |
| chr6_-_39396691 | 1.90 |
ENSMUST00000146785.8
ENSMUST00000114823.8 |
Mkrn1
|
makorin, ring finger protein, 1 |
| chr4_+_156194427 | 1.89 |
ENSMUST00000072554.13
ENSMUST00000169550.8 ENSMUST00000105576.2 |
9430015G10Rik
|
RIKEN cDNA 9430015G10 gene |
| chr5_+_137786031 | 1.88 |
ENSMUST00000035852.14
|
Zcwpw1
|
zinc finger, CW type with PWWP domain 1 |
| chr4_-_126096551 | 1.86 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chrX_+_42680037 | 1.84 |
ENSMUST00000105113.4
|
Tex13c1
|
TEX13 family member C1 |
| chr11_-_115405200 | 1.83 |
ENSMUST00000021083.7
|
Jpt1
|
Jupiter microtubule associated homolog 1 |
| chr11_-_102255999 | 1.83 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr4_+_62443606 | 1.82 |
ENSMUST00000062145.2
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr12_+_51395154 | 1.81 |
ENSMUST00000121521.2
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr19_-_45738002 | 1.80 |
ENSMUST00000070215.8
|
Npm3
|
nucleoplasmin 3 |
| chr10_-_93425553 | 1.80 |
ENSMUST00000020203.7
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr2_+_84670543 | 1.78 |
ENSMUST00000111624.8
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr17_-_80022480 | 1.78 |
ENSMUST00000234361.2
|
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr12_+_116369017 | 1.78 |
ENSMUST00000084828.5
ENSMUST00000222469.2 ENSMUST00000221114.2 ENSMUST00000221970.2 |
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr4_-_70328659 | 1.77 |
ENSMUST00000144099.8
|
Cdk5rap2
|
CDK5 regulatory subunit associated protein 2 |
| chr7_-_30251680 | 1.77 |
ENSMUST00000215288.2
ENSMUST00000108165.8 ENSMUST00000153594.3 |
Proser3
|
proline and serine rich 3 |
| chr5_-_92457862 | 1.75 |
ENSMUST00000031364.5
|
Sdad1
|
SDA1 domain containing 1 |
| chr4_-_126096376 | 1.72 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr5_-_100867520 | 1.71 |
ENSMUST00000112908.2
ENSMUST00000045617.15 |
Hpse
|
heparanase |
| chr5_-_113957318 | 1.68 |
ENSMUST00000201194.4
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr3_-_58544622 | 1.67 |
ENSMUST00000041115.7
|
Erich6
|
glutamate rich 6 |
| chr2_+_104961228 | 1.67 |
ENSMUST00000111098.8
ENSMUST00000111099.2 |
Wt1
|
Wilms tumor 1 homolog |
| chr12_+_51395047 | 1.61 |
ENSMUST00000119211.8
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr2_+_84670956 | 1.60 |
ENSMUST00000111625.2
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr17_+_27136065 | 1.55 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B |
| chr13_-_38221045 | 1.54 |
ENSMUST00000089840.5
|
Cage1
|
cancer antigen 1 |
| chr11_+_116089678 | 1.54 |
ENSMUST00000021130.7
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr5_-_92457753 | 1.52 |
ENSMUST00000201143.2
|
Sdad1
|
SDA1 domain containing 1 |
| chr17_-_45785752 | 1.50 |
ENSMUST00000233553.2
ENSMUST00000233769.2 ENSMUST00000024731.9 |
Spats1
|
spermatogenesis associated, serine-rich 1 |
| chr19_+_53588808 | 1.49 |
ENSMUST00000025930.10
|
Smc3
|
structural maintenance of chromosomes 3 |
| chr17_+_80514889 | 1.49 |
ENSMUST00000134652.2
|
Ttc39d
|
tetratricopeptide repeat domain 39D |
| chr12_-_112792971 | 1.48 |
ENSMUST00000062092.7
ENSMUST00000220899.2 |
Cdca4
|
cell division cycle associated 4 |
| chr5_+_115697526 | 1.47 |
ENSMUST00000086519.12
ENSMUST00000156359.2 ENSMUST00000152976.2 |
Rplp0
|
ribosomal protein, large, P0 |
| chr10_+_79763164 | 1.45 |
ENSMUST00000105376.2
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr2_-_30691709 | 1.44 |
ENSMUST00000050003.9
|
1700001O22Rik
|
RIKEN cDNA 1700001O22 gene |
| chr11_+_88864753 | 1.43 |
ENSMUST00000036649.8
|
Coil
|
coilin |
| chr4_+_119280002 | 1.39 |
ENSMUST00000094819.5
|
Zmynd12
|
zinc finger, MYND domain containing 12 |
| chr12_+_51394812 | 1.38 |
ENSMUST00000054308.13
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr7_+_43057611 | 1.37 |
ENSMUST00000005592.7
|
Siglecg
|
sialic acid binding Ig-like lectin G |
| chr3_-_105594865 | 1.34 |
ENSMUST00000090680.11
|
Ddx20
|
DEAD box helicase 20 |
| chr15_+_103148824 | 1.34 |
ENSMUST00000036004.16
ENSMUST00000087351.9 ENSMUST00000231141.2 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr2_-_29983618 | 1.31 |
ENSMUST00000081838.7
ENSMUST00000102865.11 |
Zdhhc12
|
zinc finger, DHHC domain containing 12 |
| chr11_-_60668067 | 1.29 |
ENSMUST00000120417.8
ENSMUST00000102668.10 ENSMUST00000117743.8 ENSMUST00000130068.3 ENSMUST00000002891.11 |
Top3a
|
topoisomerase (DNA) III alpha |
| chr2_+_150751475 | 1.29 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr14_+_30723340 | 1.29 |
ENSMUST00000168584.9
ENSMUST00000226378.2 |
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr10_+_86541675 | 1.27 |
ENSMUST00000075632.14
ENSMUST00000217747.2 ENSMUST00000061458.9 |
Ttc41
|
tetratricopeptide repeat domain 41 |
| chr3_-_19365431 | 1.24 |
ENSMUST00000099195.10
|
Pde7a
|
phosphodiesterase 7A |
| chr4_+_123798625 | 1.23 |
ENSMUST00000030400.14
|
Mycbp
|
MYC binding protein |
| chr9_-_65330231 | 1.23 |
ENSMUST00000065894.7
|
Slc51b
|
solute carrier family 51, beta subunit |
| chr10_+_98942898 | 1.22 |
ENSMUST00000020113.15
ENSMUST00000159228.8 ENSMUST00000159990.2 |
Poc1b
|
POC1 centriolar protein B |
| chr14_-_103336990 | 1.22 |
ENSMUST00000022720.15
ENSMUST00000144141.8 |
Fbxl3
|
F-box and leucine-rich repeat protein 3 |
| chr5_+_34140777 | 1.22 |
ENSMUST00000094869.12
ENSMUST00000114383.8 |
Gm1673
|
predicted gene 1673 |
| chr4_-_155859014 | 1.21 |
ENSMUST00000042196.4
|
Vwa1
|
von Willebrand factor A domain containing 1 |
| chr4_-_128856213 | 1.20 |
ENSMUST00000119354.8
ENSMUST00000106068.8 ENSMUST00000030581.10 |
Azin2
|
antizyme inhibitor 2 |
| chr12_+_111383864 | 1.19 |
ENSMUST00000220537.2
ENSMUST00000223050.2 ENSMUST00000072646.8 ENSMUST00000223431.2 ENSMUST00000221144.2 ENSMUST00000222437.2 |
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr17_+_34823236 | 1.18 |
ENSMUST00000174041.8
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chrX_-_17185555 | 1.18 |
ENSMUST00000026014.8
|
Efhc2
|
EF-hand domain (C-terminal) containing 2 |
| chr14_+_30723371 | 1.17 |
ENSMUST00000022476.9
|
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr13_-_99027544 | 1.15 |
ENSMUST00000109399.9
|
Tnpo1
|
transportin 1 |
| chr6_+_29272463 | 1.15 |
ENSMUST00000115289.2
|
Hilpda
|
hypoxia inducible lipid droplet associated |
| chr1_-_191307648 | 1.13 |
ENSMUST00000027933.11
|
Dtl
|
denticleless E3 ubiquitin protein ligase |
| chr13_+_49806542 | 1.12 |
ENSMUST00000222197.2
ENSMUST00000221083.2 ENSMUST00000223467.2 |
Nol8
|
nucleolar protein 8 |
| chr17_-_35023521 | 1.11 |
ENSMUST00000025223.9
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr9_+_59564482 | 1.09 |
ENSMUST00000216620.2
ENSMUST00000217038.2 |
Pkm
|
pyruvate kinase, muscle |
| chr2_+_28423367 | 1.08 |
ENSMUST00000113893.8
ENSMUST00000100241.10 |
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr4_-_88595161 | 1.08 |
ENSMUST00000105148.2
|
Ifna16
|
interferon alpha 16 |
| chr13_-_58506890 | 1.08 |
ENSMUST00000225388.2
|
Kif27
|
kinesin family member 27 |
| chr3_+_32763313 | 1.07 |
ENSMUST00000126144.3
|
Actl6a
|
actin-like 6A |
| chr8_-_112026854 | 1.06 |
ENSMUST00000038739.5
|
Rfwd3
|
ring finger and WD repeat domain 3 |
| chr9_+_57468217 | 1.05 |
ENSMUST00000045791.11
ENSMUST00000216986.2 |
Scamp2
|
secretory carrier membrane protein 2 |
| chr1_+_179495767 | 1.05 |
ENSMUST00000040538.10
|
Sccpdh
|
saccharopine dehydrogenase (putative) |
| chrX_+_77413242 | 1.02 |
ENSMUST00000035626.8
|
4930480E11Rik
|
RIKEN cDNA 4930480E11 gene |
| chr3_+_32762656 | 1.01 |
ENSMUST00000029214.14
|
Actl6a
|
actin-like 6A |
| chr5_+_33815892 | 1.00 |
ENSMUST00000152847.8
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr8_-_84963653 | 0.99 |
ENSMUST00000039480.7
|
Zswim4
|
zinc finger SWIM-type containing 4 |
| chr5_-_137305895 | 0.99 |
ENSMUST00000199243.5
ENSMUST00000197466.5 ENSMUST00000040873.12 |
Srrt
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr9_-_59393893 | 0.99 |
ENSMUST00000171975.8
|
Arih1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr19_+_47568041 | 0.97 |
ENSMUST00000026043.12
|
Slk
|
STE20-like kinase |
| chr7_+_16576821 | 0.96 |
ENSMUST00000168093.9
|
Prkd2
|
protein kinase D2 |
| chr2_-_120867529 | 0.95 |
ENSMUST00000102490.10
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr6_-_70769135 | 0.95 |
ENSMUST00000066134.6
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr11_-_104333059 | 0.94 |
ENSMUST00000106977.8
ENSMUST00000106972.8 |
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr3_-_82783790 | 0.94 |
ENSMUST00000048647.14
|
Rbm46
|
RNA binding motif protein 46 |
| chr10_+_13428638 | 0.93 |
ENSMUST00000019944.9
|
Adat2
|
adenosine deaminase, tRNA-specific 2 |
| chr5_+_34140877 | 0.93 |
ENSMUST00000114382.2
|
Gm1673
|
predicted gene 1673 |
| chr4_-_43578823 | 0.92 |
ENSMUST00000030189.14
|
Gba2
|
glucosidase beta 2 |
| chr12_-_113760187 | 0.92 |
ENSMUST00000192911.2
ENSMUST00000103455.3 |
Ighv2-6-8
|
immunoglobulin heavy variable 2-6-8 |
| chr7_-_27713540 | 0.92 |
ENSMUST00000180502.8
|
Zfp850
|
zinc finger protein 850 |
| chrX_+_100838256 | 0.91 |
ENSMUST00000151231.10
|
8030474K03Rik
|
RIKEN cDNA 8030474K03 gene |
| chr1_+_178233640 | 0.91 |
ENSMUST00000027775.9
|
Efcab2
|
EF-hand calcium binding domain 2 |
| chr5_-_137016355 | 0.89 |
ENSMUST00000137272.2
ENSMUST00000111090.9 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr7_-_102143980 | 0.89 |
ENSMUST00000058750.4
|
Olfr545
|
olfactory receptor 545 |
| chr7_+_140774061 | 0.88 |
ENSMUST00000084446.9
ENSMUST00000070458.11 |
Lrrc56
|
leucine rich repeat containing 56 |
| chr4_+_123798690 | 0.87 |
ENSMUST00000106202.4
|
Mycbp
|
MYC binding protein |
| chr17_-_23892798 | 0.87 |
ENSMUST00000047436.11
ENSMUST00000115490.9 ENSMUST00000095579.11 |
Thoc6
|
THO complex 6 |
| chr19_+_53131187 | 0.87 |
ENSMUST00000050096.15
ENSMUST00000237832.2 |
Add3
|
adducin 3 (gamma) |
| chr1_+_69866145 | 0.85 |
ENSMUST00000065425.12
ENSMUST00000113940.4 |
Spag16
|
sperm associated antigen 16 |
| chr3_-_32546380 | 0.84 |
ENSMUST00000164954.3
|
Kcnmb3
|
potassium large conductance calcium-activated channel, subfamily M, beta member 3 |
| chr14_-_63746541 | 0.84 |
ENSMUST00000022522.15
|
Tdh
|
L-threonine dehydrogenase |
| chr1_-_131172903 | 0.83 |
ENSMUST00000027688.15
ENSMUST00000112442.2 |
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr10_-_13428582 | 0.83 |
ENSMUST00000105541.8
|
Pex3
|
peroxisomal biogenesis factor 3 |
| chr4_-_43578636 | 0.82 |
ENSMUST00000130443.2
|
Gba2
|
glucosidase beta 2 |
| chr17_-_36291087 | 0.82 |
ENSMUST00000055454.14
|
Prr3
|
proline-rich polypeptide 3 |
| chr10_-_59057570 | 0.82 |
ENSMUST00000220156.2
ENSMUST00000165971.3 |
Septin10
|
septin 10 |
| chr5_-_30278552 | 0.82 |
ENSMUST00000198095.2
ENSMUST00000196872.2 ENSMUST00000026846.11 |
Tyms
|
thymidylate synthase |
| chr19_+_46293210 | 0.81 |
ENSMUST00000236591.2
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr1_-_36408202 | 0.80 |
ENSMUST00000185912.7
|
Kansl3
|
KAT8 regulatory NSL complex subunit 3 |
| chr2_+_172841907 | 0.78 |
ENSMUST00000029013.10
ENSMUST00000132212.2 |
Rae1
|
ribonucleic acid export 1 |
| chr17_+_14168635 | 0.78 |
ENSMUST00000088809.6
|
Gm7168
|
predicted gene 7168 |
| chr2_+_121786444 | 0.77 |
ENSMUST00000036647.13
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr1_+_131455635 | 0.77 |
ENSMUST00000068613.5
|
Fam72a
|
family with sequence similarity 72, member A |
| chr5_-_115257336 | 0.76 |
ENSMUST00000031524.11
|
Acads
|
acyl-Coenzyme A dehydrogenase, short chain |
| chr2_+_75489596 | 0.75 |
ENSMUST00000111964.8
ENSMUST00000111962.8 ENSMUST00000111961.8 ENSMUST00000164947.9 ENSMUST00000090792.11 |
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr3_-_96634752 | 0.75 |
ENSMUST00000154679.8
|
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C |
| chr3_-_96634880 | 0.75 |
ENSMUST00000029741.9
|
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C |
| chr11_-_88754543 | 0.75 |
ENSMUST00000107904.3
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr6_-_124689094 | 0.74 |
ENSMUST00000004379.8
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr9_-_20978389 | 0.73 |
ENSMUST00000115494.3
|
Zglp1
|
zinc finger, GATA-like protein 1 |
| chr11_-_116515026 | 0.73 |
ENSMUST00000103029.10
|
Rhbdf2
|
rhomboid 5 homolog 2 |
| chr3_+_151916059 | 0.72 |
ENSMUST00000166984.8
|
Fubp1
|
far upstream element (FUSE) binding protein 1 |
| chr2_+_31578537 | 0.71 |
ENSMUST00000075759.13
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr19_-_29721012 | 0.71 |
ENSMUST00000175764.9
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr7_-_24997291 | 0.71 |
ENSMUST00000148150.8
ENSMUST00000155118.2 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr9_+_99125420 | 0.71 |
ENSMUST00000185799.7
ENSMUST00000093795.10 ENSMUST00000190715.7 ENSMUST00000191335.7 ENSMUST00000190078.7 |
Cep70
|
centrosomal protein 70 |
| chr17_+_28059036 | 0.70 |
ENSMUST00000071006.9
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr1_+_91468796 | 0.70 |
ENSMUST00000188081.7
ENSMUST00000188879.2 |
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr10_-_13428855 | 0.70 |
ENSMUST00000105539.2
|
Pex3
|
peroxisomal biogenesis factor 3 |
| chr17_-_80514725 | 0.69 |
ENSMUST00000234696.2
ENSMUST00000235069.2 ENSMUST00000063417.11 |
Srsf7
|
serine and arginine-rich splicing factor 7 |
| chr17_-_65847731 | 0.69 |
ENSMUST00000233117.2
ENSMUST00000062161.7 |
Tmem232
|
transmembrane protein 232 |
| chr6_-_146479323 | 0.69 |
ENSMUST00000032427.15
|
Ints13
|
integrator complex subunit 13 |
| chr13_-_99027482 | 0.69 |
ENSMUST00000179301.8
ENSMUST00000179271.2 |
Tnpo1
|
transportin 1 |
| chr19_-_45548942 | 0.69 |
ENSMUST00000026239.7
|
Poll
|
polymerase (DNA directed), lambda |
| chr6_-_149090146 | 0.68 |
ENSMUST00000095319.10
ENSMUST00000141346.2 ENSMUST00000111535.8 |
Amn1
|
antagonist of mitotic exit network 1 |
| chr5_+_145077172 | 0.68 |
ENSMUST00000162594.8
ENSMUST00000162308.8 ENSMUST00000159018.8 ENSMUST00000160075.2 |
Bud31
|
BUD31 homolog |
| chr16_-_84532630 | 0.68 |
ENSMUST00000116584.2
|
Mrpl39
|
mitochondrial ribosomal protein L39 |
| chr17_-_65847777 | 0.67 |
ENSMUST00000086722.10
|
Tmem232
|
transmembrane protein 232 |
| chr5_-_145077048 | 0.67 |
ENSMUST00000031627.9
|
Pdap1
|
PDGFA associated protein 1 |
| chr13_+_52737118 | 0.67 |
ENSMUST00000120135.8
|
Syk
|
spleen tyrosine kinase |
| chr2_-_120867232 | 0.66 |
ENSMUST00000023987.6
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr4_+_152123772 | 0.66 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chr14_+_52962756 | 0.66 |
ENSMUST00000181483.3
|
Trav6d-3
|
T cell receptor alpha variable 6D-3 |
| chr11_-_117859997 | 0.66 |
ENSMUST00000054002.4
|
Socs3
|
suppressor of cytokine signaling 3 |
| chr12_+_100165694 | 0.66 |
ENSMUST00000110082.11
|
Calm1
|
calmodulin 1 |
| chr7_-_116042674 | 0.65 |
ENSMUST00000170430.3
ENSMUST00000206219.2 |
Pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 alpha |
| chr4_-_126096112 | 0.64 |
ENSMUST00000142125.2
ENSMUST00000106141.3 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr2_-_164498503 | 0.64 |
ENSMUST00000109335.8
|
Wfdc9
|
WAP four-disulfide core domain 9 |
| chr6_+_29272625 | 0.64 |
ENSMUST00000054445.9
|
Hilpda
|
hypoxia inducible lipid droplet associated |
| chr19_+_6097083 | 0.63 |
ENSMUST00000134667.8
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr13_-_103470937 | 0.62 |
ENSMUST00000167058.8
ENSMUST00000164111.2 |
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 1.0 | 4.9 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.9 | 2.8 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 0.9 | 2.6 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.9 | 3.5 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 0.8 | 4.1 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.8 | 2.3 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.8 | 3.8 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.7 | 3.5 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.6 | 3.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.6 | 1.8 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.6 | 1.7 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.6 | 1.7 | GO:2001076 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.4 | 1.8 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.4 | 2.2 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.4 | 1.4 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.3 | 1.7 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.3 | 0.7 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.3 | 2.0 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.3 | 4.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.3 | 2.6 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.3 | 1.9 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.3 | 1.3 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.3 | 1.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 0.9 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.3 | 2.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 1.3 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.3 | 0.8 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 3.0 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 2.7 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.2 | 0.7 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.2 | 0.9 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 1.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 0.6 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.2 | 2.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 0.6 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.2 | 1.8 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 0.7 | GO:0045401 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) cellular response to molecule of fungal origin(GO:0071226) |
| 0.2 | 0.8 | GO:0006231 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.2 | 2.0 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 1.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.4 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.1 | 3.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 3.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 1.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 1.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 3.0 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 0.4 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.1 | 0.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.5 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.7 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 4.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 2.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 2.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.0 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.7 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 1.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 4.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.6 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.8 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 0.9 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.4 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 2.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 2.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 1.7 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 1.0 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 0.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 2.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 1.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 3.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 1.1 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) |
| 0.1 | 1.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.4 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 2.0 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 2.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 1.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.5 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 | 0.8 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.1 | 0.9 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.4 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 1.0 | GO:0033129 | positive regulation of histone phosphorylation(GO:0033129) |
| 0.0 | 0.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 5.8 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 0.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 1.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.7 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 2.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.0 | 0.5 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 1.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.8 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 1.1 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 1.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 1.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.5 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 2.0 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.3 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 4.2 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.9 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 1.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.7 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.3 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 2.1 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 1.3 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.2 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.7 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 5.5 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:1904871 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.0 | GO:1904349 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) negative regulation of locomotion involved in locomotory behavior(GO:0090327) positive regulation of eating behavior(GO:1904000) positive regulation of small intestine smooth muscle contraction(GO:1904349) |
| 0.0 | 1.6 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.8 | 6.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.8 | 2.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.7 | 2.8 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.5 | 1.5 | GO:1990879 | CST complex(GO:1990879) |
| 0.4 | 1.3 | GO:0000811 | GINS complex(GO:0000811) |
| 0.4 | 4.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.4 | 1.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.3 | 1.8 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.3 | 0.9 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.3 | 1.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 1.5 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 1.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 1.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 3.8 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 0.9 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 0.6 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 2.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 1.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.2 | 2.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.9 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 2.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.1 | 0.5 | GO:0071920 | cleavage body(GO:0071920) |
| 0.1 | 3.8 | GO:0031231 | intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 3.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 3.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.9 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 3.1 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.1 | 1.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 2.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.7 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.7 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 1.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 2.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 3.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 4.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 2.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.1 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 2.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 4.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:0005662 | Prp19 complex(GO:0000974) DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 3.4 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 1.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.8 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.8 | 3.4 | GO:0072354 | histone kinase activity (H3-T3 specific)(GO:0072354) |
| 0.6 | 4.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.6 | 1.7 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.6 | 2.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.5 | 2.8 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.5 | 2.7 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.4 | 1.5 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.3 | 2.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 1.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.3 | 0.9 | GO:0016509 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 1.7 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 1.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 1.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 0.6 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 0.7 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.2 | 1.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 0.5 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.2 | 0.7 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 2.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.4 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.7 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.8 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 2.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.7 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 2.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.7 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.5 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.1 | 2.6 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 1.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 3.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.7 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.3 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.9 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 3.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 2.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 3.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.4 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.7 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 1.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 2.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 2.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 2.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 1.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 1.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 2.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 3.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 1.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 2.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 3.2 | GO:0005516 | calmodulin binding(GO:0005516) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 5.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 4.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 4.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 2.2 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 3.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 0.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 2.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 3.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.6 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.4 | 1.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 3.1 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.1 | 2.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 3.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 4.9 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 1.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.8 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 1.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 1.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 4.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 3.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 1.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 2.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 5.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 1.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 2.5 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 1.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 3.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |