Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Hivep1
Z-value: 1.31
Transcription factors associated with Hivep1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hivep1
|
ENSMUSG00000021366.9 | Hivep1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hivep1 | mm39_v1_chr13_+_42205790_42205880 | -0.37 | 1.2e-03 | Click! |
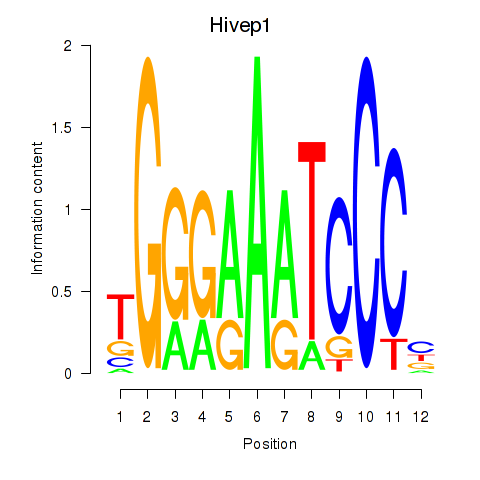
Activity profile of Hivep1 motif
Sorted Z-values of Hivep1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hivep1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_34219225 | 13.09 |
ENSMUST00000238098.2
ENSMUST00000087189.7 ENSMUST00000173075.3 ENSMUST00000172912.8 ENSMUST00000236740.2 ENSMUST00000025181.18 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr6_-_129252396 | 12.56 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr17_+_35598583 | 10.60 |
ENSMUST00000081435.5
|
H2-Q4
|
histocompatibility 2, Q region locus 4 |
| chr17_+_35482063 | 9.19 |
ENSMUST00000172503.3
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr11_+_100751151 | 9.10 |
ENSMUST00000138083.8
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr17_+_35413415 | 8.77 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr6_+_71176811 | 8.75 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr17_-_37794434 | 8.40 |
ENSMUST00000016427.11
ENSMUST00000171139.3 |
H2-M2
|
histocompatibility 2, M region locus 2 |
| chr17_+_35780977 | 8.03 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr9_-_70842090 | 7.96 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr12_-_113912416 | 7.68 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chr9_+_5308828 | 7.53 |
ENSMUST00000162846.8
ENSMUST00000027012.14 |
Casp4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr9_-_46146558 | 7.46 |
ENSMUST00000121916.8
ENSMUST00000034586.9 |
Apoc3
|
apolipoprotein C-III |
| chr1_+_192835414 | 7.35 |
ENSMUST00000076521.7
|
Irf6
|
interferon regulatory factor 6 |
| chr17_+_35481702 | 7.34 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr7_-_79492091 | 7.29 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr9_-_46146928 | 7.11 |
ENSMUST00000118649.8
|
Apoc3
|
apolipoprotein C-III |
| chr12_-_113561594 | 7.03 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr11_+_100751272 | 6.43 |
ENSMUST00000107357.4
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr12_+_85733415 | 6.43 |
ENSMUST00000040536.6
|
Batf
|
basic leucine zipper transcription factor, ATF-like |
| chr6_+_55313409 | 6.28 |
ENSMUST00000004774.4
|
Aqp1
|
aquaporin 1 |
| chr1_-_36312482 | 6.22 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr6_+_41090484 | 6.02 |
ENSMUST00000103267.3
|
Trbv12-1
|
T cell receptor beta, variable 12-1 |
| chr7_+_79836581 | 6.00 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr4_-_43031370 | 5.97 |
ENSMUST00000138030.2
ENSMUST00000136326.8 |
Stoml2
|
stomatin (Epb7.2)-like 2 |
| chr6_-_129252323 | 5.68 |
ENSMUST00000204411.2
|
Cd69
|
CD69 antigen |
| chr13_-_100382831 | 5.68 |
ENSMUST00000049789.3
|
Naip5
|
NLR family, apoptosis inhibitory protein 5 |
| chr17_-_35081456 | 5.66 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr15_+_78128990 | 5.63 |
ENSMUST00000096357.12
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr5_-_98178811 | 5.61 |
ENSMUST00000031281.14
|
Antxr2
|
anthrax toxin receptor 2 |
| chr7_-_46365108 | 5.60 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr18_-_20192535 | 5.40 |
ENSMUST00000075214.9
ENSMUST00000039247.11 |
Dsc2
|
desmocollin 2 |
| chr12_-_113823290 | 5.33 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr11_+_100750316 | 5.00 |
ENSMUST00000107356.8
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr15_+_78129040 | 4.77 |
ENSMUST00000133618.3
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr13_+_43938251 | 4.74 |
ENSMUST00000015540.4
|
Cd83
|
CD83 antigen |
| chr9_-_70841881 | 4.73 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chr14_-_55828511 | 4.72 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr18_+_84106796 | 4.72 |
ENSMUST00000235383.2
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr19_+_34268071 | 4.66 |
ENSMUST00000112472.4
ENSMUST00000235232.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr19_+_34268053 | 4.65 |
ENSMUST00000025691.13
|
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr3_+_20039775 | 4.63 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin |
| chr5_-_98178834 | 4.55 |
ENSMUST00000199088.2
|
Antxr2
|
anthrax toxin receptor 2 |
| chr5_-_144294854 | 4.46 |
ENSMUST00000055190.8
|
Baiap2l1
|
BAI1-associated protein 2-like 1 |
| chr6_+_83034825 | 4.41 |
ENSMUST00000077502.5
|
Dqx1
|
DEAQ RNA-dependent ATPase |
| chr14_+_53698556 | 4.36 |
ENSMUST00000181728.3
|
Trav7-4
|
T cell receptor alpha variable 7-4 |
| chr17_-_34109513 | 4.36 |
ENSMUST00000173386.2
ENSMUST00000114361.9 ENSMUST00000173492.9 |
Kifc1
|
kinesin family member C1 |
| chr8_+_95472218 | 4.35 |
ENSMUST00000034231.4
|
Ccl22
|
chemokine (C-C motif) ligand 22 |
| chr7_-_19363280 | 4.34 |
ENSMUST00000094762.10
ENSMUST00000049912.15 ENSMUST00000098754.5 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr12_-_55539372 | 4.27 |
ENSMUST00000021413.9
|
Nfkbia
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
| chr4_-_60455331 | 4.24 |
ENSMUST00000135953.2
|
Mup1
|
major urinary protein 1 |
| chr3_-_65865656 | 4.23 |
ENSMUST00000029416.14
|
Ccnl1
|
cyclin L1 |
| chr18_+_84106188 | 4.22 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr16_-_30086317 | 4.22 |
ENSMUST00000064856.9
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr2_-_126341757 | 4.18 |
ENSMUST00000040128.12
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr9_-_78389006 | 4.13 |
ENSMUST00000042235.15
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr15_-_76501525 | 4.10 |
ENSMUST00000230977.2
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr17_+_45866618 | 3.98 |
ENSMUST00000024742.9
ENSMUST00000233929.2 |
Nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, epsilon |
| chr11_+_98851238 | 3.94 |
ENSMUST00000107473.3
|
Rara
|
retinoic acid receptor, alpha |
| chr2_-_93292257 | 3.88 |
ENSMUST00000150508.8
|
Cd82
|
CD82 antigen |
| chr7_+_30121776 | 3.82 |
ENSMUST00000108175.8
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr5_-_92496730 | 3.79 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr4_-_57916283 | 3.74 |
ENSMUST00000063816.6
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr18_-_4352944 | 3.68 |
ENSMUST00000025078.10
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chrX_+_35375751 | 3.68 |
ENSMUST00000033418.8
|
Il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr2_-_153079828 | 3.62 |
ENSMUST00000109795.2
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr16_+_45430743 | 3.61 |
ENSMUST00000161347.9
ENSMUST00000023339.5 |
Gcsam
|
germinal center associated, signaling and motility |
| chr6_+_83011154 | 3.57 |
ENSMUST00000000707.9
ENSMUST00000101257.4 |
Loxl3
|
lysyl oxidase-like 3 |
| chr6_-_48817914 | 3.48 |
ENSMUST00000164733.4
|
Tmem176b
|
transmembrane protein 176B |
| chr11_+_75546671 | 3.41 |
ENSMUST00000108431.3
|
Myo1c
|
myosin IC |
| chr11_-_23720953 | 3.38 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr3_+_127584251 | 3.35 |
ENSMUST00000164447.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr13_-_19521337 | 3.34 |
ENSMUST00000103563.3
|
Trgv2
|
T cell receptor gamma variable 2 |
| chr3_+_127584449 | 3.30 |
ENSMUST00000171621.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr6_-_48818006 | 3.29 |
ENSMUST00000203229.3
|
Tmem176b
|
transmembrane protein 176B |
| chr11_+_97697328 | 3.22 |
ENSMUST00000153520.3
|
Lasp1
|
LIM and SH3 protein 1 |
| chr13_-_100453124 | 3.20 |
ENSMUST00000042220.3
|
Naip6
|
NLR family, apoptosis inhibitory protein 6 |
| chrX_+_84913911 | 3.13 |
ENSMUST00000113976.2
|
Tasl
|
TLR adaptor interacting with endolysosomal SLC15A4 |
| chr11_+_75546989 | 3.13 |
ENSMUST00000136935.2
|
Myo1c
|
myosin IC |
| chr6_-_48818044 | 3.09 |
ENSMUST00000101429.11
ENSMUST00000204073.3 |
Tmem176b
|
transmembrane protein 176B |
| chr14_-_51045182 | 3.06 |
ENSMUST00000227614.2
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr7_+_101032021 | 3.05 |
ENSMUST00000141083.9
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr1_-_13062873 | 2.99 |
ENSMUST00000188454.7
|
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr12_-_113625906 | 2.98 |
ENSMUST00000103448.3
|
Ighv5-9
|
immunoglobulin heavy variable 5-9 |
| chr14_+_53562089 | 2.98 |
ENSMUST00000178100.3
|
Trav7n-6
|
T cell receptor alpha variable 7N-6 |
| chr7_-_79382573 | 2.95 |
ENSMUST00000205747.2
|
Plin1
|
perilipin 1 |
| chr17_-_84495364 | 2.83 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr14_+_53007210 | 2.83 |
ENSMUST00000178768.4
|
Trav7d-4
|
T cell receptor alpha variable 7D-4 |
| chr12_-_8351954 | 2.69 |
ENSMUST00000220073.2
ENSMUST00000037313.6 |
Gdf7
|
growth differentiation factor 7 |
| chr11_-_70128678 | 2.69 |
ENSMUST00000108575.9
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr7_-_4400704 | 2.67 |
ENSMUST00000108590.4
ENSMUST00000206928.2 |
Gp6
|
glycoprotein 6 (platelet) |
| chr6_-_48818302 | 2.66 |
ENSMUST00000203355.3
ENSMUST00000166247.8 |
Tmem176b
|
transmembrane protein 176B |
| chr12_+_111383864 | 2.63 |
ENSMUST00000220537.2
ENSMUST00000223050.2 ENSMUST00000072646.8 ENSMUST00000223431.2 ENSMUST00000221144.2 ENSMUST00000222437.2 |
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr7_-_79382503 | 2.62 |
ENSMUST00000032762.14
ENSMUST00000205915.2 |
Plin1
|
perilipin 1 |
| chr2_-_69172944 | 2.54 |
ENSMUST00000102709.8
ENSMUST00000102710.10 ENSMUST00000180142.2 |
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr12_-_113542610 | 2.52 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr1_-_172418058 | 2.52 |
ENSMUST00000065679.8
|
Slamf8
|
SLAM family member 8 |
| chr14_+_53680701 | 2.48 |
ENSMUST00000177622.4
|
Trav7-3
|
T cell receptor alpha variable 7-3 |
| chr11_-_101066266 | 2.43 |
ENSMUST00000062759.4
|
Ccr10
|
chemokine (C-C motif) receptor 10 |
| chr19_+_37674029 | 2.42 |
ENSMUST00000073391.5
|
Cyp26c1
|
cytochrome P450, family 26, subfamily c, polypeptide 1 |
| chr16_-_52272828 | 2.41 |
ENSMUST00000170035.8
ENSMUST00000164728.8 ENSMUST00000168071.2 |
Alcam
|
activated leukocyte cell adhesion molecule |
| chr4_+_123176570 | 2.40 |
ENSMUST00000106243.8
ENSMUST00000106241.8 ENSMUST00000080178.13 |
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr4_+_114914607 | 2.38 |
ENSMUST00000136946.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr10_-_95251327 | 2.37 |
ENSMUST00000172070.8
ENSMUST00000150432.8 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr11_+_97697128 | 2.36 |
ENSMUST00000138919.2
|
Lasp1
|
LIM and SH3 protein 1 |
| chr3_+_122277372 | 2.33 |
ENSMUST00000197073.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr10_-_18891095 | 2.31 |
ENSMUST00000019997.11
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr7_-_79382651 | 2.29 |
ENSMUST00000205413.2
|
Plin1
|
perilipin 1 |
| chr3_+_115681788 | 2.27 |
ENSMUST00000196804.5
ENSMUST00000106505.8 ENSMUST00000043342.10 |
Dph5
|
diphthamide biosynthesis 5 |
| chr2_+_84867783 | 2.20 |
ENSMUST00000168266.8
ENSMUST00000130729.3 |
Ssrp1
|
structure specific recognition protein 1 |
| chr5_-_134776101 | 2.16 |
ENSMUST00000015138.13
|
Eln
|
elastin |
| chr6_+_42377172 | 2.16 |
ENSMUST00000057398.4
|
Tas2r143
|
taste receptor, type 2, member 143 |
| chr11_-_70128712 | 2.15 |
ENSMUST00000108577.8
ENSMUST00000108579.8 ENSMUST00000021181.13 ENSMUST00000108578.9 ENSMUST00000102569.10 |
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr17_+_43978377 | 2.14 |
ENSMUST00000233627.2
ENSMUST00000233437.2 |
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr1_+_132996237 | 2.12 |
ENSMUST00000239467.2
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr3_+_130906434 | 2.11 |
ENSMUST00000098611.4
|
Lef1
|
lymphoid enhancer binding factor 1 |
| chr2_-_118377500 | 2.10 |
ENSMUST00000125860.3
|
Bmf
|
BCL2 modifying factor |
| chr4_+_155646807 | 2.08 |
ENSMUST00000030939.14
|
Nadk
|
NAD kinase |
| chr11_+_94102255 | 2.06 |
ENSMUST00000041589.6
|
Tob1
|
transducer of ErbB-2.1 |
| chr5_+_137348423 | 2.06 |
ENSMUST00000111055.9
|
Ephb4
|
Eph receptor B4 |
| chr10_+_111342147 | 2.01 |
ENSMUST00000164773.2
|
Phlda1
|
pleckstrin homology like domain, family A, member 1 |
| chr5_+_90708962 | 1.95 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr17_+_27136065 | 1.92 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B |
| chr11_-_99176086 | 1.92 |
ENSMUST00000017255.4
|
Krt24
|
keratin 24 |
| chr7_+_30121147 | 1.89 |
ENSMUST00000108176.9
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr6_-_83030759 | 1.85 |
ENSMUST00000134606.8
|
Htra2
|
HtrA serine peptidase 2 |
| chr2_+_160730076 | 1.84 |
ENSMUST00000109457.3
|
Lpin3
|
lipin 3 |
| chr11_-_93846453 | 1.81 |
ENSMUST00000072566.5
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr17_+_43978280 | 1.81 |
ENSMUST00000170988.2
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr5_+_137348363 | 1.80 |
ENSMUST00000061244.15
|
Ephb4
|
Eph receptor B4 |
| chr12_-_28685849 | 1.78 |
ENSMUST00000221871.2
|
Rps7
|
ribosomal protein S7 |
| chr6_-_67468831 | 1.77 |
ENSMUST00000118364.2
|
Il23r
|
interleukin 23 receptor |
| chr15_+_102379621 | 1.73 |
ENSMUST00000229918.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr10_-_95251145 | 1.71 |
ENSMUST00000119917.2
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr17_+_31896781 | 1.65 |
ENSMUST00000228716.3
ENSMUST00000019192.8 ENSMUST00000228089.3 |
Cryaa
|
crystallin, alpha A |
| chr15_+_102379503 | 1.64 |
ENSMUST00000229222.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr5_-_138169509 | 1.64 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr4_+_151012375 | 1.58 |
ENSMUST00000139826.8
ENSMUST00000116257.8 |
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr2_+_160730019 | 1.56 |
ENSMUST00000109455.9
ENSMUST00000040872.13 |
Lpin3
|
lipin 3 |
| chr14_+_53628092 | 1.55 |
ENSMUST00000103636.4
|
Trav7-2
|
T cell receptor alpha variable 7-2 |
| chr9_-_32255604 | 1.55 |
ENSMUST00000034533.7
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr2_-_103133503 | 1.54 |
ENSMUST00000111176.9
|
Ehf
|
ets homologous factor |
| chr4_-_40239700 | 1.48 |
ENSMUST00000142055.2
|
Ddx58
|
DEAD/H box helicase 58 |
| chr6_-_97464761 | 1.47 |
ENSMUST00000032146.14
|
Frmd4b
|
FERM domain containing 4B |
| chr8_-_123349473 | 1.43 |
ENSMUST00000212966.2
ENSMUST00000093059.3 |
Pabpn1l
|
poly(A)binding protein nuclear 1-like |
| chr4_-_40239778 | 1.40 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr2_+_32611067 | 1.40 |
ENSMUST00000074248.11
|
Sh2d3c
|
SH2 domain containing 3C |
| chr9_-_32255556 | 1.37 |
ENSMUST00000214223.2
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr4_-_133695264 | 1.36 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr14_+_51045298 | 1.36 |
ENSMUST00000036126.7
|
Parp2
|
poly (ADP-ribose) polymerase family, member 2 |
| chr11_-_70120503 | 1.34 |
ENSMUST00000153449.2
ENSMUST00000000326.12 |
Bcl6b
|
B cell CLL/lymphoma 6, member B |
| chr13_-_120488547 | 1.32 |
ENSMUST00000177659.2
|
Gm36079
|
predicted gene, 36079 |
| chr3_+_88857929 | 1.32 |
ENSMUST00000186583.7
|
Ash1l
|
ASH1 like histone lysine methyltransferase |
| chr2_+_61408875 | 1.32 |
ENSMUST00000112502.8
ENSMUST00000078074.9 |
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr2_-_45003270 | 1.30 |
ENSMUST00000202935.4
ENSMUST00000068415.11 ENSMUST00000127520.8 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr3_+_90200470 | 1.30 |
ENSMUST00000199754.5
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr12_-_28685913 | 1.30 |
ENSMUST00000074267.5
|
Rps7
|
ribosomal protein S7 |
| chr7_-_99276310 | 1.29 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr9_-_32255533 | 1.23 |
ENSMUST00000216033.2
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr2_-_113588983 | 1.18 |
ENSMUST00000099575.4
|
Grem1
|
gremlin 1, DAN family BMP antagonist |
| chr17_-_83939316 | 1.16 |
ENSMUST00000234613.2
ENSMUST00000051482.2 |
Kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr2_-_103133524 | 1.15 |
ENSMUST00000090475.10
|
Ehf
|
ets homologous factor |
| chr4_-_43836517 | 1.13 |
ENSMUST00000215442.2
|
Olfr157
|
olfactory receptor 157 |
| chrX_-_133501677 | 1.10 |
ENSMUST00000239113.2
|
Gla
|
galactosidase, alpha |
| chr2_-_5068807 | 1.09 |
ENSMUST00000114996.8
|
Optn
|
optineurin |
| chr12_-_113945961 | 1.06 |
ENSMUST00000103466.2
|
Ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr6_+_113284098 | 1.06 |
ENSMUST00000113122.8
ENSMUST00000204198.3 ENSMUST00000113121.8 ENSMUST00000113119.8 ENSMUST00000113117.4 ENSMUST00000204626.3 ENSMUST00000203577.2 |
Brpf1
|
bromodomain and PHD finger containing, 1 |
| chr10_-_127358300 | 1.04 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chrX_-_135680890 | 0.99 |
ENSMUST00000018739.5
|
Glra4
|
glycine receptor, alpha 4 subunit |
| chr2_+_125876566 | 0.95 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
| chr8_+_91555449 | 0.94 |
ENSMUST00000109614.9
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr14_+_55798517 | 0.91 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr9_-_57672375 | 0.89 |
ENSMUST00000215233.2
|
Clk3
|
CDC-like kinase 3 |
| chr6_+_48425034 | 0.85 |
ENSMUST00000212740.2
ENSMUST00000169350.9 ENSMUST00000043676.12 |
Sspo
|
SCO-spondin |
| chr5_-_138169253 | 0.84 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr14_+_55798362 | 0.83 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr11_+_51153941 | 0.83 |
ENSMUST00000093132.13
ENSMUST00000109113.8 |
Clk4
|
CDC like kinase 4 |
| chr2_-_5068743 | 0.74 |
ENSMUST00000027986.5
|
Optn
|
optineurin |
| chr14_+_52981706 | 0.71 |
ENSMUST00000179789.4
|
Trav7d-3
|
T cell receptor alpha variable 7D-3 |
| chr19_-_12093187 | 0.69 |
ENSMUST00000208391.3
ENSMUST00000214103.2 |
Olfr1428
|
olfactory receptor 1428 |
| chr2_+_118730823 | 0.67 |
ENSMUST00000151162.2
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chrX_-_161747552 | 0.67 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G |
| chr14_+_20979466 | 0.59 |
ENSMUST00000022369.9
|
Vcl
|
vinculin |
| chr16_-_58710581 | 0.58 |
ENSMUST00000058564.4
|
Olfr178
|
olfactory receptor 178 |
| chr10_-_127358231 | 0.57 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr11_-_100628979 | 0.57 |
ENSMUST00000155500.2
ENSMUST00000107364.8 ENSMUST00000019317.12 |
Rab5c
|
RAB5C, member RAS oncogene family |
| chr6_-_21852508 | 0.56 |
ENSMUST00000031678.10
|
Tspan12
|
tetraspanin 12 |
| chr14_+_55797934 | 0.54 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr9_-_110818679 | 0.51 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chr11_+_69920542 | 0.51 |
ENSMUST00000232266.2
ENSMUST00000132597.5 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr6_-_21851827 | 0.49 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr13_-_120496867 | 0.48 |
ENSMUST00000178618.2
|
Gm21188
|
predicted gene, 21188 |
| chr3_-_129834788 | 0.46 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr12_+_3956808 | 0.45 |
ENSMUST00000172913.2
|
Dnmt3a
|
DNA methyltransferase 3A |
| chr6_-_83513184 | 0.45 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr3_-_132655954 | 0.45 |
ENSMUST00000042744.16
ENSMUST00000117811.8 |
Npnt
|
nephronectin |
| chr17_-_28736483 | 0.44 |
ENSMUST00000114792.8
ENSMUST00000177939.8 |
Fkbp5
|
FK506 binding protein 5 |
| chr5_-_37981919 | 0.42 |
ENSMUST00000063116.10
|
Msx1
|
msh homeobox 1 |
| chr4_+_115747862 | 0.42 |
ENSMUST00000132221.3
ENSMUST00000165938.2 |
6430628N08Rik
|
RIKEN cDNA 6430628N08 gene |
| chr1_+_186947934 | 0.42 |
ENSMUST00000160471.8
|
Gpatch2
|
G patch domain containing 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 29.6 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 5.1 | 20.5 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 3.6 | 14.6 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 3.1 | 9.3 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 2.5 | 12.7 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 2.1 | 6.3 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 1.5 | 6.0 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 1.3 | 12.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 1.3 | 4.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 1.3 | 3.9 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 1.2 | 3.6 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 1.2 | 3.6 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 1.2 | 16.4 | GO:0070269 | pyroptosis(GO:0070269) |
| 1.1 | 6.6 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 1.1 | 4.4 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 1.0 | 4.0 | GO:0032329 | serine transport(GO:0032329) |
| 1.0 | 12.5 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.9 | 3.8 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.9 | 3.7 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.9 | 3.7 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.8 | 4.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.8 | 6.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.8 | 2.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.7 | 2.1 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.7 | 6.9 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.6 | 3.9 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.6 | 1.9 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.6 | 5.4 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.6 | 2.9 | GO:0009597 | detection of virus(GO:0009597) |
| 0.6 | 9.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.5 | 26.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.5 | 2.7 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.5 | 1.6 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.5 | 2.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.5 | 5.7 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) |
| 0.5 | 3.6 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.5 | 6.4 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.5 | 1.4 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.5 | 2.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.4 | 1.3 | GO:1903699 | tarsal gland development(GO:1903699) |
| 0.4 | 4.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.4 | 4.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.4 | 2.5 | GO:0046618 | drug export(GO:0046618) |
| 0.4 | 4.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.3 | 7.3 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.3 | 2.4 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.3 | 5.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 1.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.3 | 1.8 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.3 | 2.8 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.3 | 3.5 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.2 | 10.4 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.2 | 7.3 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.2 | 1.7 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.2 | 5.6 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 2.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 2.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.6 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.2 | 18.2 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.2 | 3.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 0.6 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.2 | 4.7 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.2 | 2.1 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.2 | 1.3 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 4.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 4.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 25.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 10.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 2.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 3.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 4.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.8 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.1 | 5.6 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.1 | 1.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 4.6 | GO:0046688 | copper ion transport(GO:0006825) response to copper ion(GO:0046688) |
| 0.1 | 7.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.5 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.4 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.1 | 1.0 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 6.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 6.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 0.3 | GO:1904550 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.1 | 0.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 3.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 2.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 3.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 2.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 2.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.2 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.1 | 1.4 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.5 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 1.3 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.0 | 0.7 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 1.7 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.5 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 1.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.6 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 4.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0060158 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 2.4 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 1.3 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 5.7 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 2.7 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 3.4 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 48.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 2.3 | 16.4 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 1.6 | 6.3 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 1.3 | 14.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.3 | 6.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 1.0 | 9.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.8 | 8.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.8 | 2.4 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.7 | 7.3 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.6 | 10.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.6 | 7.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.6 | 18.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 2.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 4.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 8.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 2.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 25.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 1.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 2.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 2.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 4.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 5.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 4.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 2.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 39.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 9.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 8.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 2.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 8.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 4.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 2.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 8.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 4.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 2.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 2.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 4.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 12.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 3.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 4.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 2.8 | 48.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 2.5 | 12.7 | GO:0035478 | chylomicron binding(GO:0035478) |
| 2.3 | 9.3 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 2.2 | 8.9 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 1.4 | 4.2 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 1.3 | 4.0 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 1.0 | 6.3 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 1.0 | 8.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.9 | 3.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.8 | 10.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.7 | 5.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.7 | 4.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.6 | 3.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.6 | 7.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.6 | 6.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.6 | 3.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.6 | 4.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.5 | 1.6 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.5 | 1.5 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.4 | 2.4 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.4 | 4.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 4.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 1.8 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.3 | 3.1 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.3 | 1.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 2.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.3 | 1.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.3 | 7.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.3 | 1.8 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.3 | 6.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 3.9 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.2 | 3.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 1.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.2 | 10.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 5.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 1.8 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 25.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 8.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 2.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 3.9 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 4.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 1.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 3.0 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 4.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 21.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 2.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 1.0 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 4.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.4 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 4.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 2.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 4.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.1 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 3.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 3.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 2.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 4.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 4.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 2.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 3.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 4.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.5 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 2.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 4.4 | GO:0070035 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) ATP-dependent helicase activity(GO:0008026) RNA-dependent ATPase activity(GO:0008186) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 4.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 4.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 2.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 7.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 4.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 2.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 9.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 8.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 5.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 2.7 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 2.3 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 4.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.6 | 20.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.6 | 9.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.4 | 10.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 8.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.3 | 5.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 3.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 5.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 3.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 4.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 7.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 4.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 1.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 4.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 6.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 3.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 7.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 7.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 27.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.9 | 20.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.5 | 9.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.4 | 6.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 5.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 2.9 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.3 | 4.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.3 | 2.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 7.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 4.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 8.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 10.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 8.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 10.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 2.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 11.5 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 4.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 4.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 7.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 3.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 4.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 4.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 7.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 8.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 2.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |