Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
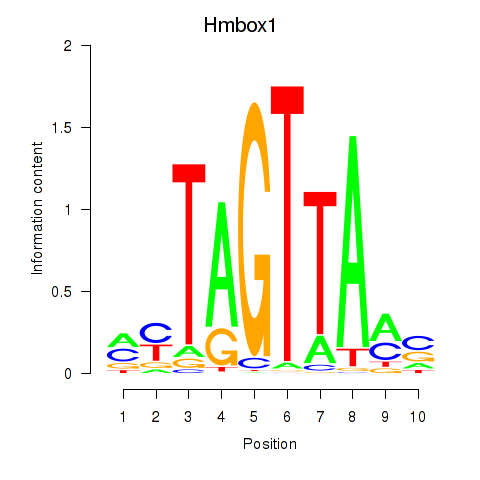
Results for Hmbox1
Z-value: 1.15
Transcription factors associated with Hmbox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmbox1
|
ENSMUSG00000021972.15 | Hmbox1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmbox1 | mm39_v1_chr14_-_65187287_65187335 | -0.31 | 8.9e-03 | Click! |
Activity profile of Hmbox1 motif
Sorted Z-values of Hmbox1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmbox1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_28752050 | 8.81 |
ENSMUST00000029240.14
|
Slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr1_+_21310821 | 7.83 |
ENSMUST00000121676.8
ENSMUST00000124990.3 |
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr1_+_21310803 | 7.65 |
ENSMUST00000027067.15
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr4_+_117109204 | 7.48 |
ENSMUST00000125943.8
ENSMUST00000106434.8 |
Tmem53
|
transmembrane protein 53 |
| chr4_+_100336003 | 7.15 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr11_+_22958338 | 7.04 |
ENSMUST00000172602.9
|
Fam161a
|
family with sequence similarity 161, member A |
| chr11_+_22958418 | 7.03 |
ENSMUST00000109557.10
|
Fam161a
|
family with sequence similarity 161, member A |
| chr6_-_113411718 | 6.58 |
ENSMUST00000113091.8
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chr1_+_88128323 | 6.47 |
ENSMUST00000049289.9
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr4_+_98919183 | 5.92 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr11_-_88742285 | 5.38 |
ENSMUST00000107903.8
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr13_+_4484305 | 5.02 |
ENSMUST00000021630.15
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr19_+_32573182 | 4.95 |
ENSMUST00000235594.2
|
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr5_+_87148697 | 4.85 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr8_-_45715049 | 4.85 |
ENSMUST00000034064.5
|
F11
|
coagulation factor XI |
| chr2_+_70339157 | 4.78 |
ENSMUST00000100041.9
|
Erich2
|
glutamate rich 2 |
| chr2_+_70339175 | 4.73 |
ENSMUST00000134607.8
|
Erich2
|
glutamate rich 2 |
| chr8_-_106863423 | 4.40 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr8_-_106863521 | 4.34 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr7_-_44711075 | 4.34 |
ENSMUST00000007981.9
ENSMUST00000210500.2 ENSMUST00000210493.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr19_-_28945194 | 4.31 |
ENSMUST00000162110.8
|
Spata6l
|
spermatogenesis associated 6 like |
| chr2_+_34764408 | 4.30 |
ENSMUST00000113068.9
ENSMUST00000047447.13 |
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr18_-_10706701 | 4.20 |
ENSMUST00000002549.9
ENSMUST00000117726.9 ENSMUST00000117828.9 |
Abhd3
|
abhydrolase domain containing 3 |
| chr6_-_24168082 | 4.11 |
ENSMUST00000031713.9
|
Slc13a1
|
solute carrier family 13 (sodium/sulfate symporters), member 1 |
| chr18_-_3281089 | 4.09 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr11_-_94398162 | 3.82 |
ENSMUST00000040692.9
|
Mycbpap
|
MYCBP associated protein |
| chr2_+_173561208 | 3.82 |
ENSMUST00000073081.6
|
1700010B08Rik
|
RIKEN cDNA 1700010B08 gene |
| chr18_-_3280999 | 3.80 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr7_-_44711130 | 3.75 |
ENSMUST00000211337.2
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr1_-_185849448 | 3.69 |
ENSMUST00000045388.8
|
Lyplal1
|
lysophospholipase-like 1 |
| chr13_-_73826124 | 3.67 |
ENSMUST00000022105.15
ENSMUST00000109680.10 ENSMUST00000221026.2 ENSMUST00000109679.4 |
Slc6a18
|
solute carrier family 6 (neurotransmitter transporter), member 18 |
| chr11_-_78313043 | 3.61 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr4_+_52596266 | 3.60 |
ENSMUST00000029995.6
|
Toporsl
|
topoisomerase I binding, arginine/serine-rich like |
| chr2_+_34764496 | 3.59 |
ENSMUST00000028228.6
|
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr4_+_128887017 | 3.49 |
ENSMUST00000030583.13
ENSMUST00000102604.11 |
Ak2
|
adenylate kinase 2 |
| chr10_-_39009910 | 3.47 |
ENSMUST00000135785.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr19_+_6155804 | 3.44 |
ENSMUST00000044451.4
|
Naaladl1
|
N-acetylated alpha-linked acidic dipeptidase-like 1 |
| chr16_-_43836681 | 3.38 |
ENSMUST00000036174.10
|
Gramd1c
|
GRAM domain containing 1C |
| chr10_-_39009844 | 3.28 |
ENSMUST00000134279.8
ENSMUST00000139743.8 ENSMUST00000149949.8 ENSMUST00000124941.8 ENSMUST00000125042.8 ENSMUST00000063204.9 |
Fam229b
|
family with sequence similarity 229, member B |
| chr19_+_39275518 | 3.24 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr16_-_21814190 | 3.22 |
ENSMUST00000231766.2
ENSMUST00000074230.12 |
Liph
|
lipase, member H |
| chr2_+_70339832 | 3.13 |
ENSMUST00000153121.2
|
Erich2
|
glutamate rich 2 |
| chrX_+_162873183 | 3.13 |
ENSMUST00000015545.10
|
Cltrn
|
collectrin, amino acid transport regulator |
| chr15_+_54975713 | 3.12 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr11_-_94932158 | 3.08 |
ENSMUST00000038431.8
|
Pdk2
|
pyruvate dehydrogenase kinase, isoenzyme 2 |
| chr9_+_106391771 | 3.05 |
ENSMUST00000085113.5
|
Iqcf5
|
IQ motif containing F5 |
| chrM_+_5319 | 3.02 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr7_+_134870237 | 2.98 |
ENSMUST00000210697.2
ENSMUST00000097983.5 |
Nps
|
neuropeptide S |
| chr4_+_20042045 | 2.95 |
ENSMUST00000098242.4
|
Ggh
|
gamma-glutamyl hydrolase |
| chr11_+_31822211 | 2.84 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr13_+_22220000 | 2.80 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12 |
| chr6_+_30610973 | 2.76 |
ENSMUST00000062758.11
|
Cpa5
|
carboxypeptidase A5 |
| chr1_+_163979384 | 2.75 |
ENSMUST00000086040.6
|
F5
|
coagulation factor V |
| chr13_-_73826108 | 2.72 |
ENSMUST00000222029.2
ENSMUST00000223074.2 ENSMUST00000220650.2 ENSMUST00000221987.2 |
Slc6a18
|
solute carrier family 6 (neurotransmitter transporter), member 18 |
| chr8_+_46387647 | 2.70 |
ENSMUST00000095326.10
|
Ccdc110
|
coiled-coil domain containing 110 |
| chr4_+_51216645 | 2.65 |
ENSMUST00000166749.2
ENSMUST00000156384.4 |
Cylc2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr7_-_130748035 | 2.65 |
ENSMUST00000070980.4
|
4933402N03Rik
|
RIKEN cDNA 4933402N03 gene |
| chr6_+_30611028 | 2.62 |
ENSMUST00000115138.8
|
Cpa5
|
carboxypeptidase A5 |
| chr16_-_21814289 | 2.62 |
ENSMUST00000060673.8
|
Liph
|
lipase, member H |
| chr4_-_58987094 | 2.59 |
ENSMUST00000030069.7
|
Ptgr1
|
prostaglandin reductase 1 |
| chr9_-_71075939 | 2.59 |
ENSMUST00000113570.8
|
Aqp9
|
aquaporin 9 |
| chr6_+_5390386 | 2.58 |
ENSMUST00000183358.2
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr13_-_35108293 | 2.56 |
ENSMUST00000223834.2
|
Fam217a
|
family with sequence similarity 217, member A |
| chr10_+_100428212 | 2.55 |
ENSMUST00000187119.7
ENSMUST00000188736.7 ENSMUST00000191336.7 |
1700017N19Rik
|
RIKEN cDNA 1700017N19 gene |
| chr9_-_78275430 | 2.54 |
ENSMUST00000071991.6
|
Dppa5a
|
developmental pluripotency associated 5A |
| chr6_+_29853745 | 2.49 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr3_-_79749949 | 2.46 |
ENSMUST00000029568.7
|
Tmem144
|
transmembrane protein 144 |
| chr15_+_54975814 | 2.43 |
ENSMUST00000100660.11
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr10_+_100428246 | 2.38 |
ENSMUST00000041162.13
ENSMUST00000190386.7 ENSMUST00000190708.7 |
1700017N19Rik
|
RIKEN cDNA 1700017N19 gene |
| chr1_+_31261889 | 2.36 |
ENSMUST00000027230.3
|
Dnaaf6
|
dynein axonemal assembly factor 6 |
| chr15_-_100576715 | 2.33 |
ENSMUST00000229869.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr4_-_63090355 | 2.29 |
ENSMUST00000156618.9
ENSMUST00000030042.3 |
Kif12
|
kinesin family member 12 |
| chr3_+_132335704 | 2.27 |
ENSMUST00000212594.2
|
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr7_-_39062584 | 2.25 |
ENSMUST00000108017.2
|
Gm5114
|
predicted gene 5114 |
| chr13_+_68011442 | 2.24 |
ENSMUST00000078471.7
|
BC048507
|
cDNA sequence BC048507 |
| chr17_+_41245569 | 2.23 |
ENSMUST00000169611.4
|
Mmut
|
methylmalonyl-Coenzyme A mutase |
| chr12_+_52144511 | 2.23 |
ENSMUST00000040090.16
|
Nubpl
|
nucleotide binding protein-like |
| chr8_-_94739469 | 2.21 |
ENSMUST00000053766.14
|
Amfr
|
autocrine motility factor receptor |
| chr11_+_51654511 | 2.21 |
ENSMUST00000020653.6
|
Sar1b
|
secretion associated Ras related GTPase 1B |
| chr13_-_96678844 | 2.19 |
ENSMUST00000223475.2
|
Polk
|
polymerase (DNA directed), kappa |
| chr5_+_7354130 | 2.17 |
ENSMUST00000160634.2
ENSMUST00000159546.2 |
Tex47
|
testis expressed 47 |
| chr1_+_88154727 | 2.14 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr7_-_38227617 | 2.13 |
ENSMUST00000079759.6
|
Gm5591
|
predicted gene 5591 |
| chr12_+_36364136 | 2.11 |
ENSMUST00000041407.7
|
Sostdc1
|
sclerostin domain containing 1 |
| chr2_+_3771709 | 2.07 |
ENSMUST00000177037.2
|
Fam107b
|
family with sequence similarity 107, member B |
| chr11_+_11414256 | 2.06 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chrX_+_42680037 | 2.02 |
ENSMUST00000105113.4
|
Tex13c1
|
TEX13 family member C1 |
| chr2_+_85551751 | 1.95 |
ENSMUST00000055517.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr13_-_3943556 | 1.94 |
ENSMUST00000099946.6
|
Net1
|
neuroepithelial cell transforming gene 1 |
| chrX_+_156482116 | 1.85 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chrX_+_143239577 | 1.83 |
ENSMUST00000155206.8
|
Trpc5os
|
transient receptor potential cation channel, subfamily C, member 5, opposite strand |
| chr19_+_47854249 | 1.81 |
ENSMUST00000238084.2
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr7_-_25516041 | 1.80 |
ENSMUST00000043314.10
|
Cyp2s1
|
cytochrome P450, family 2, subfamily s, polypeptide 1 |
| chr9_-_49397508 | 1.74 |
ENSMUST00000055096.5
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr12_-_81579614 | 1.73 |
ENSMUST00000169158.2
ENSMUST00000164431.2 ENSMUST00000163402.8 ENSMUST00000166664.2 ENSMUST00000164386.8 |
Synj2bp
Gm20498
|
synaptojanin 2 binding protein predicted gene 20498 |
| chr9_-_119812042 | 1.72 |
ENSMUST00000214058.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr19_-_4977997 | 1.69 |
ENSMUST00000236758.2
|
Dpp3
|
dipeptidylpeptidase 3 |
| chr11_+_73068063 | 1.68 |
ENSMUST00000108477.2
|
Tax1bp3
|
Tax1 (human T cell leukemia virus type I) binding protein 3 |
| chr14_+_54413727 | 1.64 |
ENSMUST00000103701.2
|
Traj41
|
T cell receptor alpha joining 41 |
| chr13_-_3943433 | 1.64 |
ENSMUST00000222504.2
|
Net1
|
neuroepithelial cell transforming gene 1 |
| chr11_+_73067909 | 1.64 |
ENSMUST00000040687.12
|
Tax1bp3
|
Tax1 (human T cell leukemia virus type I) binding protein 3 |
| chr19_+_45003304 | 1.59 |
ENSMUST00000039016.14
|
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chr12_+_65012564 | 1.57 |
ENSMUST00000066296.9
ENSMUST00000223166.2 |
Togaram1
|
TOG array regulator of axonemal microtubules 1 |
| chr4_-_133066594 | 1.50 |
ENSMUST00000043305.14
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr5_+_7354113 | 1.49 |
ENSMUST00000088796.3
|
Tex47
|
testis expressed 47 |
| chr4_-_133066549 | 1.49 |
ENSMUST00000105906.2
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr13_+_55359598 | 1.47 |
ENSMUST00000224918.2
|
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr7_+_40682143 | 1.46 |
ENSMUST00000164422.2
|
Gm4884
|
predicted gene 4884 |
| chr10_-_95159933 | 1.46 |
ENSMUST00000053594.7
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr19_-_32080496 | 1.41 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr13_+_55359866 | 1.40 |
ENSMUST00000224693.2
|
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr1_+_52884172 | 1.40 |
ENSMUST00000159352.8
ENSMUST00000044478.7 |
Hibch
|
3-hydroxyisobutyryl-Coenzyme A hydrolase |
| chr1_-_31261678 | 1.36 |
ENSMUST00000187892.8
ENSMUST00000233331.2 |
4931428L18Rik
|
RIKEN cDNA 4931428L18 gene |
| chr2_+_20742115 | 1.21 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr5_+_136721938 | 1.20 |
ENSMUST00000196068.5
ENSMUST00000005611.10 |
Myl10
|
myosin, light chain 10, regulatory |
| chrX_+_156481906 | 1.19 |
ENSMUST00000136141.2
ENSMUST00000190091.7 |
Smpx
|
small muscle protein, X-linked |
| chrX_-_65347749 | 1.16 |
ENSMUST00000033525.3
|
4930447F04Rik
|
RIKEN cDNA 4930447F04 gene |
| chr10_+_20024203 | 1.14 |
ENSMUST00000020173.16
|
Map7
|
microtubule-associated protein 7 |
| chr15_+_99291491 | 1.09 |
ENSMUST00000159531.3
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr4_-_141450710 | 1.00 |
ENSMUST00000102484.5
ENSMUST00000177592.2 |
Ddi2
|
DNA-damage inducible protein 2 |
| chr6_+_43150886 | 0.99 |
ENSMUST00000059512.5
|
Olfr13
|
olfactory receptor 13 |
| chr3_+_95496239 | 0.95 |
ENSMUST00000177390.8
ENSMUST00000060323.12 ENSMUST00000098861.11 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr5_+_62968993 | 0.93 |
ENSMUST00000238474.2
|
Dthd1
|
death domain containing 1 |
| chr11_+_77576981 | 0.92 |
ENSMUST00000100802.11
ENSMUST00000181023.2 |
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr10_+_129072073 | 0.91 |
ENSMUST00000203248.3
|
Olfr774
|
olfactory receptor 774 |
| chr14_-_36628263 | 0.90 |
ENSMUST00000183007.2
|
Ccser2
|
coiled-coil serine rich 2 |
| chr14_-_70561231 | 0.90 |
ENSMUST00000151011.8
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr11_+_75541324 | 0.80 |
ENSMUST00000102505.10
|
Myo1c
|
myosin IC |
| chr6_-_97156032 | 0.79 |
ENSMUST00000095664.6
|
Tmf1
|
TATA element modulatory factor 1 |
| chr11_-_73067828 | 0.78 |
ENSMUST00000108480.2
ENSMUST00000054952.4 |
Emc6
|
ER membrane protein complex subunit 6 |
| chr1_+_155916655 | 0.76 |
ENSMUST00000065648.15
ENSMUST00000097526.3 |
Tor1aip2
|
torsin A interacting protein 2 |
| chr13_+_56757389 | 0.76 |
ENSMUST00000045173.10
|
Tgfbi
|
transforming growth factor, beta induced |
| chr9_+_119170360 | 0.75 |
ENSMUST00000039784.12
|
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chrX_+_141464722 | 0.73 |
ENSMUST00000112896.9
|
Tmem164
|
transmembrane protein 164 |
| chr2_+_146854916 | 0.72 |
ENSMUST00000028921.6
|
Xrn2
|
5'-3' exoribonuclease 2 |
| chr17_+_35055962 | 0.71 |
ENSMUST00000173874.8
ENSMUST00000180043.8 ENSMUST00000046244.15 |
Dxo
|
decapping exoribonuclease |
| chr3_+_95496270 | 0.68 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr3_-_95125002 | 0.65 |
ENSMUST00000107209.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr13_+_47347301 | 0.63 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr2_+_119039456 | 0.61 |
ENSMUST00000102519.5
|
Zfyve19
|
zinc finger, FYVE domain containing 19 |
| chr3_+_144824325 | 0.61 |
ENSMUST00000098538.9
ENSMUST00000106192.9 ENSMUST00000098539.7 ENSMUST00000029920.15 |
Odf2l
|
outer dense fiber of sperm tails 2-like |
| chr3_-_57202301 | 0.60 |
ENSMUST00000171384.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr5_-_143963413 | 0.58 |
ENSMUST00000031622.13
|
Ocm
|
oncomodulin |
| chr10_-_12743915 | 0.57 |
ENSMUST00000219584.2
|
Utrn
|
utrophin |
| chr2_+_32253692 | 0.56 |
ENSMUST00000113331.8
ENSMUST00000113338.9 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr1_+_24216691 | 0.56 |
ENSMUST00000054588.15
|
Col9a1
|
collagen, type IX, alpha 1 |
| chrX_+_20982059 | 0.54 |
ENSMUST00000072593.9
|
Ssxa1
|
synovial sarcoma, X member A1 |
| chr3_+_144824644 | 0.54 |
ENSMUST00000199124.5
|
Odf2l
|
outer dense fiber of sperm tails 2-like |
| chr16_+_3408906 | 0.53 |
ENSMUST00000216259.2
|
Olfr161
|
olfactory receptor 161 |
| chrX_-_110440860 | 0.52 |
ENSMUST00000128819.8
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr17_+_74645936 | 0.51 |
ENSMUST00000224711.2
ENSMUST00000024869.8 ENSMUST00000233611.2 |
Spast
|
spastin |
| chr10_-_52071340 | 0.49 |
ENSMUST00000020045.10
|
Ros1
|
Ros1 proto-oncogene |
| chr4_+_116414855 | 0.48 |
ENSMUST00000030460.15
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr3_-_64049349 | 0.48 |
ENSMUST00000177151.9
|
Vmn2r2
|
vomeronasal 2, receptor 2 |
| chr10_-_128885867 | 0.47 |
ENSMUST00000216460.2
|
Olfr765
|
olfactory receptor 765 |
| chr4_+_116415251 | 0.45 |
ENSMUST00000106475.2
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr3_+_108561223 | 0.44 |
ENSMUST00000106609.8
|
Clcc1
|
chloride channel CLIC-like 1 |
| chr10_-_129710624 | 0.42 |
ENSMUST00000081367.2
|
Olfr814
|
olfactory receptor 814 |
| chr19_+_45003635 | 0.41 |
ENSMUST00000179108.9
|
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chr3_+_108561294 | 0.40 |
ENSMUST00000106613.2
|
Clcc1
|
chloride channel CLIC-like 1 |
| chr5_-_143963461 | 0.40 |
ENSMUST00000110702.2
|
Ocm
|
oncomodulin |
| chr10_-_128918779 | 0.39 |
ENSMUST00000213579.2
|
Olfr767
|
olfactory receptor 767 |
| chr2_-_12424212 | 0.39 |
ENSMUST00000124603.8
ENSMUST00000129993.3 ENSMUST00000028105.13 |
Mindy3
|
MINDY lysine 48 deubiquitinase 3 |
| chr3_-_104960264 | 0.38 |
ENSMUST00000098763.7
ENSMUST00000197437.5 |
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr8_-_106074585 | 0.38 |
ENSMUST00000014922.5
|
Fhod1
|
formin homology 2 domain containing 1 |
| chr15_+_100320028 | 0.36 |
ENSMUST00000132119.2
|
Gm5475
|
predicted gene 5475 |
| chr3_+_108561247 | 0.36 |
ENSMUST00000124384.8
ENSMUST00000029483.15 |
Clcc1
|
chloride channel CLIC-like 1 |
| chr1_+_173093568 | 0.35 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418 |
| chr5_+_104318542 | 0.34 |
ENSMUST00000112771.2
|
Dspp
|
dentin sialophosphoprotein |
| chr1_-_131025562 | 0.33 |
ENSMUST00000016672.11
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chr4_+_152423075 | 0.31 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr7_-_47677345 | 0.31 |
ENSMUST00000094390.3
|
Mrgprx1
|
MAS-related GPR, member X1 |
| chr17_-_31877703 | 0.24 |
ENSMUST00000236475.2
ENSMUST00000166526.9 ENSMUST00000014684.6 |
U2af1
|
U2 small nuclear ribonucleoprotein auxiliary factor (U2AF) 1 |
| chr8_+_110806390 | 0.23 |
ENSMUST00000212754.2
ENSMUST00000058804.10 |
Zfp612
|
zinc finger protein 612 |
| chr10_-_40133558 | 0.22 |
ENSMUST00000216847.2
ENSMUST00000213628.2 ENSMUST00000217537.2 ENSMUST00000019982.9 |
Gtf3c6
|
general transcription factor IIIC, polypeptide 6, alpha |
| chr1_+_192856044 | 0.22 |
ENSMUST00000193307.2
|
A130010J15Rik
|
RIKEN cDNA A130010J15 gene |
| chr12_+_103524690 | 0.21 |
ENSMUST00000187155.7
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr14_-_55101505 | 0.21 |
ENSMUST00000142283.4
|
Homez
|
homeodomain leucine zipper-encoding gene |
| chr5_+_88117307 | 0.20 |
ENSMUST00000007601.4
ENSMUST00000187738.2 |
2310003L06Rik
Gm28434
|
RIKEN cDNA 2310003L06 gene predicted gene 28434 |
| chr11_+_49321254 | 0.20 |
ENSMUST00000203369.2
|
Olfr1389
|
olfactory receptor 1389 |
| chr4_-_155170738 | 0.19 |
ENSMUST00000030914.4
|
Rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr2_+_85838122 | 0.19 |
ENSMUST00000062166.2
|
Olfr1032
|
olfactory receptor 1032 |
| chr1_-_69726384 | 0.19 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr10_-_52071052 | 0.17 |
ENSMUST00000218452.2
|
Ros1
|
Ros1 proto-oncogene |
| chr11_+_101358990 | 0.17 |
ENSMUST00000001347.7
|
Rnd2
|
Rho family GTPase 2 |
| chr16_+_35803794 | 0.17 |
ENSMUST00000173555.8
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr3_+_94350622 | 0.16 |
ENSMUST00000029786.14
ENSMUST00000196143.2 |
Mrpl9
|
mitochondrial ribosomal protein L9 |
| chr19_+_45003670 | 0.13 |
ENSMUST00000236377.2
|
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chr1_-_131207279 | 0.11 |
ENSMUST00000062108.10
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr11_-_4654303 | 0.10 |
ENSMUST00000058407.6
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr10_+_4296806 | 0.10 |
ENSMUST00000216139.3
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr3_+_93227047 | 0.07 |
ENSMUST00000090856.10
ENSMUST00000093774.4 |
Hrnr
|
hornerin |
| chr15_+_25774070 | 0.07 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr18_+_35695736 | 0.07 |
ENSMUST00000235851.2
ENSMUST00000235581.2 |
Matr3
|
matrin 3 |
| chr11_+_96909559 | 0.03 |
ENSMUST00000107622.2
|
Sp6
|
trans-acting transcription factor 6 |
| chr1_-_171476495 | 0.02 |
ENSMUST00000194791.2
ENSMUST00000192024.6 |
Slamf7
|
SLAM family member 7 |
| chr14_-_79461316 | 0.02 |
ENSMUST00000040802.5
|
Zfp957
|
zinc finger protein 957 |
| chr1_-_144427302 | 0.02 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr13_+_49658249 | 0.01 |
ENSMUST00000051504.8
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr14_+_27344385 | 0.01 |
ENSMUST00000210135.2
ENSMUST00000090302.6 ENSMUST00000211087.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 1.8 | 8.8 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 1.7 | 5.0 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.2 | 5.0 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.0 | 5.9 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 1.0 | 4.8 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.8 | 3.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.7 | 3.0 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.6 | 2.6 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.6 | 3.7 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.6 | 5.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.6 | 3.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.5 | 6.5 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 2.1 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.5 | 3.0 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.3 | 2.3 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.3 | 2.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.3 | 0.8 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.3 | 6.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 2.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.3 | 1.8 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.2 | 2.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 5.5 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 1.1 | GO:0031437 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.2 | 2.6 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 2.9 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.2 | 2.2 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.6 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.2 | 1.0 | GO:1902336 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.2 | 3.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 4.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 8.7 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.2 | 3.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 5.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 3.0 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 3.3 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 1.7 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.1 | 0.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 9.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 2.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.3 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 6.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.7 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 1.6 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.8 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.6 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 1.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 5.9 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 4.2 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 2.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.2 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.0 | 0.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.7 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 9.5 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 1.6 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 2.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.5 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 1.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.6 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.7 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 2.1 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 5.8 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 2.6 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 1.4 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.3 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.9 | 7.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.6 | 3.5 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.4 | 3.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.4 | 2.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 5.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.3 | 14.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 0.8 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.2 | 3.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 2.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.8 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.1 | 14.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 2.8 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 2.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 7.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 1.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 2.9 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 3.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 4.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 6.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 37.7 | GO:0070062 | extracellular exosome(GO:0070062) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 8.8 | GO:0055056 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 1.7 | 5.0 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.2 | 5.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.8 | 3.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.7 | 4.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.6 | 2.6 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.5 | 3.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.5 | 5.9 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.5 | 2.6 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.4 | 2.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.4 | 2.9 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.4 | 3.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.4 | 5.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.4 | 2.5 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 17.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 4.8 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.3 | 11.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 0.7 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.2 | 3.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 0.7 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.2 | 5.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 4.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 2.1 | GO:0098821 | BMP binding(GO:0036122) BMP receptor activity(GO:0098821) |
| 0.2 | 6.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.2 | 3.8 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.2 | 1.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 1.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 3.0 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 2.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 3.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 10.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 3.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.9 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 1.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 2.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 2.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 5.8 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 2.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 10.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.5 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.7 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 1.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.4 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 7.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 10.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 10.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 3.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 3.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 1.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 3.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 8.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 5.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 5.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 6.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 2.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.5 | 7.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 5.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 17.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 6.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 3.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 4.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 2.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 2.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 3.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 5.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 9.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 3.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |