Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Hmga1
Z-value: 1.15
Transcription factors associated with Hmga1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmga1
|
ENSMUSG00000046711.17 | Hmga1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmga1 | mm39_v1_chr17_+_27775471_27775609 | 0.36 | 2.0e-03 | Click! |
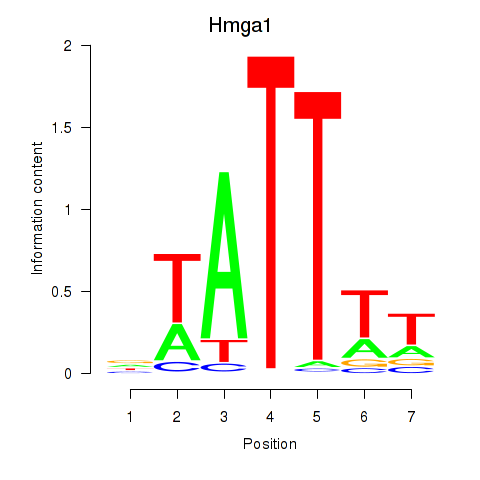
Activity profile of Hmga1 motif
Sorted Z-values of Hmga1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmga1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_7494108 | 11.25 |
ENSMUST00000193330.2
|
Pkia
|
protein kinase inhibitor, alpha |
| chr17_-_52133594 | 6.50 |
ENSMUST00000129667.8
ENSMUST00000169480.8 ENSMUST00000148559.2 |
Satb1
|
special AT-rich sequence binding protein 1 |
| chr9_-_54381409 | 6.28 |
ENSMUST00000127880.8
|
Dmxl2
|
Dmx-like 2 |
| chr8_-_65489834 | 5.59 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr8_-_65489791 | 5.40 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr4_-_102883905 | 5.27 |
ENSMUST00000084382.6
ENSMUST00000106869.3 |
Insl5
|
insulin-like 5 |
| chr3_+_84859453 | 5.09 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr4_+_101365144 | 5.04 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr7_-_84021655 | 4.80 |
ENSMUST00000208392.2
ENSMUST00000208232.2 ENSMUST00000208863.2 |
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr2_+_14393127 | 4.72 |
ENSMUST00000114731.8
ENSMUST00000082290.8 |
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr1_+_17215581 | 4.71 |
ENSMUST00000026879.8
|
Gdap1
|
ganglioside-induced differentiation-associated-protein 1 |
| chrX_+_165127688 | 4.68 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr2_-_110591909 | 4.40 |
ENSMUST00000140777.2
|
Ano3
|
anoctamin 3 |
| chr5_-_103247920 | 3.77 |
ENSMUST00000112848.8
|
Mapk10
|
mitogen-activated protein kinase 10 |
| chr2_-_73490746 | 3.76 |
ENSMUST00000102677.11
|
Chn1
|
chimerin 1 |
| chr13_-_48746836 | 3.75 |
ENSMUST00000238995.2
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr13_-_54914366 | 3.72 |
ENSMUST00000036825.14
|
Sncb
|
synuclein, beta |
| chr12_+_103524690 | 3.67 |
ENSMUST00000187155.7
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr8_+_94537910 | 3.64 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr1_-_157084252 | 3.61 |
ENSMUST00000134543.8
|
Rasal2
|
RAS protein activator like 2 |
| chr14_+_74878280 | 3.60 |
ENSMUST00000036653.5
|
Htr2a
|
5-hydroxytryptamine (serotonin) receptor 2A |
| chr19_+_45433899 | 3.58 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr2_-_17395765 | 3.45 |
ENSMUST00000177966.2
|
Nebl
|
nebulette |
| chr18_+_37822865 | 3.44 |
ENSMUST00000195112.2
|
Pcdhgb2
|
protocadherin gamma subfamily B, 2 |
| chr12_+_117621644 | 3.41 |
ENSMUST00000222105.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr18_+_37453427 | 3.31 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr13_+_83721696 | 3.24 |
ENSMUST00000197146.5
ENSMUST00000185052.6 ENSMUST00000195984.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr2_+_112097087 | 3.17 |
ENSMUST00000110987.9
ENSMUST00000028549.14 |
Slc12a6
|
solute carrier family 12, member 6 |
| chr4_-_63414188 | 3.16 |
ENSMUST00000063650.10
ENSMUST00000102867.8 ENSMUST00000107393.8 ENSMUST00000084510.8 ENSMUST00000095038.8 ENSMUST00000119294.8 ENSMUST00000095037.2 ENSMUST00000063672.10 |
Whrn
|
whirlin |
| chr18_+_37533899 | 3.05 |
ENSMUST00000057228.2
|
Pcdhb9
|
protocadherin beta 9 |
| chrX_+_135723420 | 3.05 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr5_-_5315968 | 3.03 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr2_-_58990967 | 3.01 |
ENSMUST00000226455.2
ENSMUST00000077687.6 |
Ccdc148
|
coiled-coil domain containing 148 |
| chr3_-_117153802 | 3.00 |
ENSMUST00000197743.2
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr7_-_59654849 | 2.95 |
ENSMUST00000059305.17
|
Snrpn
|
small nuclear ribonucleoprotein N |
| chr10_-_30647836 | 2.94 |
ENSMUST00000215926.2
ENSMUST00000213836.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr13_+_83723255 | 2.92 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr11_+_108573428 | 2.92 |
ENSMUST00000106718.10
ENSMUST00000106715.8 ENSMUST00000106724.10 |
Cep112
|
centrosomal protein 112 |
| chr2_+_87696836 | 2.89 |
ENSMUST00000213308.3
|
Olfr1152
|
olfactory receptor 1152 |
| chr1_+_179928709 | 2.83 |
ENSMUST00000133890.8
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr1_-_80643024 | 2.83 |
ENSMUST00000187774.7
|
Dock10
|
dedicator of cytokinesis 10 |
| chr14_+_26616514 | 2.82 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr16_-_36605286 | 2.80 |
ENSMUST00000168279.2
ENSMUST00000164579.9 ENSMUST00000023616.11 |
Slc15a2
|
solute carrier family 15 (H+/peptide transporter), member 2 |
| chr6_+_41515152 | 2.80 |
ENSMUST00000103291.2
ENSMUST00000192856.6 |
Trbc1
|
T cell receptor beta, constant region 1 |
| chr11_-_28534260 | 2.75 |
ENSMUST00000093253.10
ENSMUST00000109502.9 ENSMUST00000042534.15 |
Ccdc85a
|
coiled-coil domain containing 85A |
| chr14_-_101846459 | 2.70 |
ENSMUST00000161991.8
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chr18_+_37840092 | 2.69 |
ENSMUST00000195823.2
|
Pcdhga6
|
protocadherin gamma subfamily A, 6 |
| chr2_+_51928017 | 2.62 |
ENSMUST00000065927.6
|
Tnfaip6
|
tumor necrosis factor alpha induced protein 6 |
| chr2_-_59778560 | 2.60 |
ENSMUST00000153136.2
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr18_+_52748978 | 2.60 |
ENSMUST00000072666.4
ENSMUST00000209270.2 |
Zfp474
|
zinc finger protein 474 |
| chrX_+_94942639 | 2.58 |
ENSMUST00000082183.8
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr9_+_113641615 | 2.57 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr5_+_25964985 | 2.57 |
ENSMUST00000128727.8
ENSMUST00000088244.6 |
Actr3b
|
ARP3 actin-related protein 3B |
| chrX_+_162694397 | 2.56 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr2_+_112114906 | 2.48 |
ENSMUST00000053666.8
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr10_+_34173426 | 2.42 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chr7_+_108610032 | 2.41 |
ENSMUST00000033341.12
|
Tub
|
tubby bipartite transcription factor |
| chr5_+_35156454 | 2.41 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr5_-_121660134 | 2.39 |
ENSMUST00000100757.6
|
Adam1a
|
a disintegrin and metallopeptidase domain 1a |
| chr7_+_113365235 | 2.36 |
ENSMUST00000046687.16
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein |
| chr3_+_20011405 | 2.35 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr18_-_35841435 | 2.30 |
ENSMUST00000236738.2
ENSMUST00000237995.2 |
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr3_-_59127571 | 2.30 |
ENSMUST00000199675.2
ENSMUST00000170388.6 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr18_+_69726431 | 2.29 |
ENSMUST00000201091.4
ENSMUST00000201037.4 ENSMUST00000114977.5 |
Tcf4
|
transcription factor 4 |
| chr6_+_57679621 | 2.29 |
ENSMUST00000050077.15
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr18_+_37100672 | 2.26 |
ENSMUST00000193777.6
ENSMUST00000193389.2 |
Pcdha6
|
protocadherin alpha 6 |
| chr3_+_93349637 | 2.26 |
ENSMUST00000064257.6
|
Tchh
|
trichohyalin |
| chr5_+_20433169 | 2.24 |
ENSMUST00000197553.5
ENSMUST00000208219.2 |
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr3_+_14545751 | 2.24 |
ENSMUST00000037321.8
ENSMUST00000120484.8 ENSMUST00000120801.2 |
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr6_+_86826470 | 2.19 |
ENSMUST00000089519.13
ENSMUST00000204414.3 |
Aak1
|
AP2 associated kinase 1 |
| chr19_+_26600820 | 2.16 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr18_+_37637317 | 2.13 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr18_+_37869950 | 2.12 |
ENSMUST00000091935.7
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr15_+_41694317 | 2.12 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr16_+_91022300 | 2.11 |
ENSMUST00000035608.10
|
Olig2
|
oligodendrocyte transcription factor 2 |
| chr2_+_65760477 | 2.11 |
ENSMUST00000176109.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr8_-_68658694 | 2.10 |
ENSMUST00000212960.2
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_-_70999547 | 2.10 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chr1_-_163552693 | 2.09 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr10_+_70276604 | 2.07 |
ENSMUST00000173042.9
ENSMUST00000062883.7 |
Fam13c
|
family with sequence similarity 13, member C |
| chr6_+_17743581 | 2.01 |
ENSMUST00000000674.13
ENSMUST00000077080.9 |
St7
|
suppression of tumorigenicity 7 |
| chr7_-_139162706 | 1.99 |
ENSMUST00000106095.3
|
Nkx6-2
|
NK6 homeobox 2 |
| chr1_+_152830720 | 1.98 |
ENSMUST00000043313.15
ENSMUST00000186621.2 |
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr10_-_88440869 | 1.98 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr14_-_101846551 | 1.96 |
ENSMUST00000100340.4
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chr10_-_38998272 | 1.93 |
ENSMUST00000136546.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr3_+_20011201 | 1.93 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr18_-_22983794 | 1.93 |
ENSMUST00000092015.11
ENSMUST00000069215.13 |
Nol4
|
nucleolar protein 4 |
| chr6_+_57679455 | 1.91 |
ENSMUST00000072954.8
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr18_+_37880027 | 1.90 |
ENSMUST00000193404.2
|
Pcdhga10
|
protocadherin gamma subfamily A, 10 |
| chr12_+_49429574 | 1.88 |
ENSMUST00000179669.3
|
Foxg1
|
forkhead box G1 |
| chr12_+_29584560 | 1.88 |
ENSMUST00000021009.10
|
Myt1l
|
myelin transcription factor 1-like |
| chr1_-_77491683 | 1.87 |
ENSMUST00000186930.2
ENSMUST00000027451.13 ENSMUST00000188797.7 |
Epha4
|
Eph receptor A4 |
| chr1_+_179936757 | 1.87 |
ENSMUST00000143176.8
ENSMUST00000135056.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_+_14393245 | 1.86 |
ENSMUST00000133258.2
|
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr18_+_37085673 | 1.85 |
ENSMUST00000192512.6
ENSMUST00000192295.2 ENSMUST00000115661.5 |
Pcdha4
Gm42416
|
protocadherin alpha 4 predicted gene, 42416 |
| chr7_-_66915756 | 1.85 |
ENSMUST00000207715.2
|
Mef2a
|
myocyte enhancer factor 2A |
| chr10_-_33500583 | 1.85 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr10_+_87926932 | 1.81 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr15_+_16728842 | 1.79 |
ENSMUST00000228307.2
|
Cdh9
|
cadherin 9 |
| chr18_+_37610858 | 1.79 |
ENSMUST00000051442.7
|
Pcdhb16
|
protocadherin beta 16 |
| chr9_-_45896663 | 1.76 |
ENSMUST00000214179.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr15_+_65682066 | 1.75 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr7_-_103320398 | 1.71 |
ENSMUST00000062144.4
|
Olfr624
|
olfactory receptor 624 |
| chr3_-_66204228 | 1.71 |
ENSMUST00000029419.8
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr18_-_50701793 | 1.70 |
ENSMUST00000056460.4
|
Pudp
|
pseudouridine 5'-phosphatase |
| chr7_-_105230395 | 1.70 |
ENSMUST00000188726.2
ENSMUST00000188440.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr17_+_70829144 | 1.69 |
ENSMUST00000140728.8
|
Dlgap1
|
DLG associated protein 1 |
| chr19_-_13126896 | 1.69 |
ENSMUST00000213493.2
|
Olfr1459
|
olfactory receptor 1459 |
| chr1_-_28819331 | 1.68 |
ENSMUST00000059937.5
|
Gm597
|
predicted gene 597 |
| chr14_-_31503869 | 1.68 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr18_+_37079133 | 1.65 |
ENSMUST00000192503.2
|
Pcdha3
|
protocadherin alpha 3 |
| chr3_-_151899470 | 1.65 |
ENSMUST00000050073.13
|
Dnajb4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr5_+_14564932 | 1.65 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr18_+_37794819 | 1.64 |
ENSMUST00000194888.2
ENSMUST00000194190.2 |
Pcdhga1
|
protocadherin gamma subfamily A, 1 |
| chrX_-_58613428 | 1.64 |
ENSMUST00000119833.8
ENSMUST00000131319.8 |
Fgf13
|
fibroblast growth factor 13 |
| chr6_-_119925387 | 1.64 |
ENSMUST00000162541.8
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr8_+_47439948 | 1.63 |
ENSMUST00000119686.2
|
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr12_+_52746158 | 1.61 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr12_-_107969673 | 1.61 |
ENSMUST00000109887.8
ENSMUST00000109891.3 |
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr10_+_123099945 | 1.60 |
ENSMUST00000238972.2
ENSMUST00000050756.8 |
Tafa2
|
TAFA chemokine like family member 2 |
| chr4_+_123095297 | 1.60 |
ENSMUST00000068262.6
|
Nt5c1a
|
5'-nucleotidase, cytosolic IA |
| chr18_+_37143758 | 1.58 |
ENSMUST00000115657.10
ENSMUST00000192447.6 |
Pcdha11
|
protocadherin alpha 11 |
| chr8_-_37200051 | 1.58 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr11_-_41891111 | 1.58 |
ENSMUST00000109290.2
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chrX_+_92698469 | 1.55 |
ENSMUST00000113933.9
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr19_-_45580258 | 1.54 |
ENSMUST00000160003.8
ENSMUST00000162879.8 |
Fbxw4
|
F-box and WD-40 domain protein 4 |
| chr10_+_97482946 | 1.54 |
ENSMUST00000105285.4
|
Epyc
|
epiphycan |
| chr15_-_103123711 | 1.53 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chr10_-_108846816 | 1.52 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chr15_+_28203872 | 1.50 |
ENSMUST00000067048.8
|
Dnah5
|
dynein, axonemal, heavy chain 5 |
| chr6_-_112924205 | 1.49 |
ENSMUST00000088373.11
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr15_+_39522905 | 1.49 |
ENSMUST00000226410.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr19_-_18978463 | 1.48 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
| chr15_+_91722524 | 1.47 |
ENSMUST00000109276.8
ENSMUST00000088555.10 ENSMUST00000100293.9 ENSMUST00000126508.8 ENSMUST00000239545.1 |
Smgc
ENSMUSG00000118670.1
|
submandibular gland protein C mucin 19 |
| chr1_-_138103021 | 1.45 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr6_+_41112064 | 1.45 |
ENSMUST00000103272.4
|
Trbv14
|
T cell receptor beta, variable 14 |
| chr6_-_29164981 | 1.44 |
ENSMUST00000007993.16
|
Rbm28
|
RNA binding motif protein 28 |
| chr10_-_25076008 | 1.43 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr11_+_88964667 | 1.43 |
ENSMUST00000100619.11
|
Gm525
|
predicted gene 525 |
| chr13_+_21679387 | 1.42 |
ENSMUST00000104942.2
|
AK157302
|
cDNA sequence AK157302 |
| chrX_-_133652140 | 1.41 |
ENSMUST00000052431.12
|
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr6_+_41092928 | 1.39 |
ENSMUST00000194399.2
|
Trbv13-1
|
T cell receptor beta, variable 13-1 |
| chr7_-_112946481 | 1.39 |
ENSMUST00000117577.8
|
Btbd10
|
BTB (POZ) domain containing 10 |
| chr13_+_110063364 | 1.39 |
ENSMUST00000117420.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr17_-_78991691 | 1.38 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr16_-_50151350 | 1.37 |
ENSMUST00000114488.8
|
Bbx
|
bobby sox HMG box containing |
| chr2_+_177969456 | 1.36 |
ENSMUST00000133267.3
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr9_+_74959259 | 1.36 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr18_-_43192483 | 1.35 |
ENSMUST00000025377.14
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr18_+_37544717 | 1.34 |
ENSMUST00000051126.4
|
Pcdhb10
|
protocadherin beta 10 |
| chr12_+_73984427 | 1.34 |
ENSMUST00000221833.2
|
Snapc1l
|
small nuclear RNA activating complex, polypeptide 1 like |
| chr14_-_50479161 | 1.34 |
ENSMUST00000214388.2
|
Olfr731
|
olfactory receptor 731 |
| chr10_+_70276473 | 1.33 |
ENSMUST00000105436.9
|
Fam13c
|
family with sequence similarity 13, member C |
| chr1_-_131441962 | 1.33 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr18_-_7273578 | 1.33 |
ENSMUST00000234281.2
ENSMUST00000234196.2 |
Odad2
|
outer dynein arm docking complex subunit 2 |
| chr13_+_49806542 | 1.31 |
ENSMUST00000222197.2
ENSMUST00000221083.2 ENSMUST00000223467.2 |
Nol8
|
nucleolar protein 8 |
| chr10_+_90412114 | 1.30 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_-_112923715 | 1.30 |
ENSMUST00000113169.9
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr13_+_104424359 | 1.29 |
ENSMUST00000065766.7
|
Adamts6
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 6 |
| chr17_+_44263890 | 1.29 |
ENSMUST00000177857.9
ENSMUST00000044792.6 |
Rcan2
|
regulator of calcineurin 2 |
| chr4_-_136626073 | 1.29 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr6_+_103674695 | 1.26 |
ENSMUST00000205098.2
|
Chl1
|
cell adhesion molecule L1-like |
| chr16_-_4698148 | 1.25 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr17_+_44264130 | 1.25 |
ENSMUST00000229240.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr15_+_91722458 | 1.25 |
ENSMUST00000109277.8
|
Smgc
|
submandibular gland protein C |
| chr2_-_79959802 | 1.25 |
ENSMUST00000102653.8
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr10_-_30647881 | 1.24 |
ENSMUST00000215740.2
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr14_+_99337311 | 1.24 |
ENSMUST00000022650.9
|
Pibf1
|
progesterone immunomodulatory binding factor 1 |
| chrX_-_133652080 | 1.23 |
ENSMUST00000113194.8
|
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr12_+_110452143 | 1.23 |
ENSMUST00000221715.2
ENSMUST00000109832.3 |
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr11_+_29668563 | 1.20 |
ENSMUST00000060992.6
|
Rtn4
|
reticulon 4 |
| chrX_+_111513971 | 1.18 |
ENSMUST00000071814.13
|
Zfp711
|
zinc finger protein 711 |
| chr14_+_26414422 | 1.17 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr1_+_55445033 | 1.15 |
ENSMUST00000042986.10
|
Plcl1
|
phospholipase C-like 1 |
| chr16_-_19132814 | 1.15 |
ENSMUST00000216157.2
|
Olfr164
|
olfactory receptor 164 |
| chr1_+_128069716 | 1.14 |
ENSMUST00000187557.2
|
R3hdm1
|
R3H domain containing 1 |
| chr2_-_178049375 | 1.14 |
ENSMUST00000081134.10
|
Sycp2
|
synaptonemal complex protein 2 |
| chr1_-_160986880 | 1.14 |
ENSMUST00000135643.8
ENSMUST00000178511.3 |
Tex50
|
testis expressed 50 |
| chr9_+_111011327 | 1.11 |
ENSMUST00000216430.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr14_-_70945434 | 1.11 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr17_-_79076487 | 1.10 |
ENSMUST00000233850.2
|
Heatr5b
|
HEAT repeat containing 5B |
| chrX_-_17437801 | 1.10 |
ENSMUST00000177213.8
|
Fundc1
|
FUN14 domain containing 1 |
| chr4_+_100926170 | 1.09 |
ENSMUST00000106955.2
ENSMUST00000038463.15 |
Raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr3_-_102871440 | 1.08 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr1_-_138102972 | 1.06 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr2_+_107120934 | 1.05 |
ENSMUST00000037012.3
|
Kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr18_+_37063237 | 1.05 |
ENSMUST00000193839.6
ENSMUST00000070797.7 |
Pcdha1
|
protocadherin alpha 1 |
| chr8_+_106154266 | 1.05 |
ENSMUST00000067305.8
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr14_+_96118660 | 1.03 |
ENSMUST00000228913.2
ENSMUST00000045892.3 |
Spertl
|
spermatid associated like |
| chr1_-_126758369 | 1.03 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr5_-_122492223 | 1.02 |
ENSMUST00000117263.8
ENSMUST00000049009.7 |
Rad9b
|
RAD9 checkpoint clamp component B |
| chr1_+_179938904 | 1.02 |
ENSMUST00000145181.2
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr6_-_23650297 | 1.02 |
ENSMUST00000063548.4
|
Rnf133
|
ring finger protein 133 |
| chr14_+_43951187 | 1.02 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chr4_-_96673423 | 1.01 |
ENSMUST00000107071.2
|
Gm12695
|
predicted gene 12695 |
| chrX_-_100103249 | 1.01 |
ENSMUST00000113718.8
ENSMUST00000113716.3 |
Tex11
|
testis expressed gene 11 |
| chr6_-_119173699 | 0.99 |
ENSMUST00000239204.2
|
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr12_-_99529767 | 0.99 |
ENSMUST00000176928.3
ENSMUST00000223484.2 |
Foxn3
|
forkhead box N3 |
| chr11_+_120123727 | 0.99 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 1.0 | 4.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 1.0 | 5.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.9 | 3.7 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.9 | 3.6 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.8 | 5.0 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.8 | 2.5 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.7 | 2.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.7 | 0.7 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.7 | 1.3 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.6 | 2.5 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.6 | 1.9 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.6 | 1.9 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.6 | 1.8 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.6 | 4.7 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.6 | 2.3 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.5 | 4.4 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.5 | 1.6 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.5 | 3.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.5 | 11.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.5 | 6.2 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.5 | 3.0 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.5 | 2.4 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.5 | 5.6 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.4 | 6.3 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.4 | 2.8 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.4 | 1.9 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.3 | 6.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.3 | 2.0 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.3 | 2.0 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.3 | 1.6 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.3 | 6.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 4.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.3 | 1.0 | GO:1900063 | mitochondrial membrane fission(GO:0090149) regulation of peroxisome organization(GO:1900063) |
| 0.3 | 5.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 1.5 | GO:0006311 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.3 | 1.1 | GO:0051325 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.3 | 1.7 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.3 | 6.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.3 | 2.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 2.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 1.7 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 1.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.2 | 1.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.2 | 0.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.2 | 0.4 | GO:0002358 | B cell homeostatic proliferation(GO:0002358) |
| 0.2 | 0.9 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 3.2 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.2 | 0.6 | GO:1902071 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.2 | 0.7 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.2 | 1.6 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 1.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 1.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 1.5 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 3.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 1.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 5.7 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.2 | 5.7 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.2 | 2.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 2.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 0.8 | GO:1902774 | regulation of SNARE complex assembly(GO:0035542) late endosome to lysosome transport(GO:1902774) |
| 0.2 | 2.0 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.2 | 0.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 0.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 | 0.9 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.2 | 2.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 1.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 4.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.0 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.1 | 0.4 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.1 | 1.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 1.7 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 3.0 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 5.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.7 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 1.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.5 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 1.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 2.5 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.8 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 1.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.6 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.1 | 1.9 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.3 | GO:1904633 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.1 | 1.3 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 9.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 4.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 1.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 2.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.9 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.9 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.6 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 1.0 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 3.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.8 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 3.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 1.7 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 1.1 | GO:0071554 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.1 | 0.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.7 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 2.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 2.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.4 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.4 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.3 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 2.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 3.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.8 | GO:0035435 | cellular phosphate ion homeostasis(GO:0030643) phosphate ion transmembrane transport(GO:0035435) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 1.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 1.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 2.6 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 0.4 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.3 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 0.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 1.0 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 10.6 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 2.1 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.3 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.7 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.0 | 0.1 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 2.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 2.8 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.1 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.2 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 3.4 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 2.1 | GO:0032109 | positive regulation of macroautophagy(GO:0016239) positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.0 | 1.9 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 1.1 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.5 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 2.3 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.7 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.9 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 4.0 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.5 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.8 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0048338 | Mullerian duct regression(GO:0001880) neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.0 | 0.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 1.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 2.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0051904 | establishment of melanosome localization(GO:0032401) melanosome transport(GO:0032402) pigment granule transport(GO:0051904) establishment of pigment granule localization(GO:0051905) |
| 0.0 | 1.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.0 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 2.0 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 2.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 3.6 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 1.3 | 5.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.9 | 2.6 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.6 | 2.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.6 | 3.0 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.5 | 1.6 | GO:0044317 | rod spherule(GO:0044317) |
| 0.4 | 2.1 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.3 | 1.0 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 0.2 | 3.0 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.2 | 1.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 3.0 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 7.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 1.0 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 3.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 0.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 0.8 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.2 | 3.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 1.7 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.2 | 2.8 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 0.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 1.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 4.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.5 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 2.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 5.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 2.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.7 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.3 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.1 | 4.2 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 2.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.0 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 1.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 8.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 5.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 4.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 5.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 2.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 3.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 3.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 3.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 3.8 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.9 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 3.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 4.0 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 5.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 4.7 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 1.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 4.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.4 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.5 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 4.8 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 17.8 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 4.3 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.9 | 2.8 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.8 | 5.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.8 | 2.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.8 | 3.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 11.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.5 | 3.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.5 | 5.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 1.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 2.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 4.8 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 3.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.3 | 2.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.3 | 4.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 1.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 2.0 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.3 | 0.9 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.3 | 2.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 1.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 3.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.2 | 0.9 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 6.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 0.9 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 4.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 3.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.8 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 0.6 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.2 | 1.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.2 | 2.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 2.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 0.7 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.2 | 0.7 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.2 | 2.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 1.6 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 6.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 4.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 2.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 3.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 1.2 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.9 | GO:0050897 | retinoic acid-responsive element binding(GO:0044323) cobalt ion binding(GO:0050897) |
| 0.1 | 2.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 2.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 2.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 3.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 6.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 2.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 12.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 9.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 2.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 11.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 3.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 4.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 4.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 13.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 2.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 3.6 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 2.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 4.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 2.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.9 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 1.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.0 | 1.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.7 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 2.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 2.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 1.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.6 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 1.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 2.2 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 2.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 2.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 4.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.1 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.8 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 4.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 3.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.9 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 6.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.7 | GO:0016791 | phosphatase activity(GO:0016791) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 4.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 8.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 3.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 6.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 2.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 3.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 4.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 5.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 2.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 7.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 11.9 | REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | Genes involved in MAPK targets/ Nuclear events mediated by MAP kinases |
| 0.2 | 2.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 1.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 7.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 5.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 3.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 4.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 4.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 8.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 5.1 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 9.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 4.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |