Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
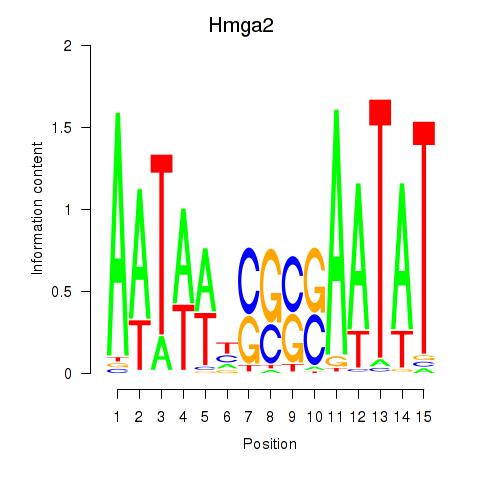
Results for Hmga2
Z-value: 0.94
Transcription factors associated with Hmga2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmga2
|
ENSMUSG00000056758.15 | Hmga2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmga2 | mm39_v1_chr10_-_120312374_120312388 | -0.53 | 1.8e-06 | Click! |
Activity profile of Hmga2 motif
Sorted Z-values of Hmga2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmga2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_87288177 | 13.55 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr5_-_87240405 | 11.91 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr5_-_87572060 | 8.26 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr19_+_40078132 | 7.92 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr6_-_142418801 | 7.57 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
| chr10_-_93375832 | 7.36 |
ENSMUST00000016034.3
|
Amdhd1
|
amidohydrolase domain containing 1 |
| chr8_-_85620537 | 6.01 |
ENSMUST00000003907.14
ENSMUST00000109745.8 ENSMUST00000142748.2 |
Gcdh
|
glutaryl-Coenzyme A dehydrogenase |
| chr5_+_90608751 | 5.53 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr2_+_163389068 | 5.47 |
ENSMUST00000109411.8
ENSMUST00000018094.13 |
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr15_-_82291372 | 5.10 |
ENSMUST00000230198.2
ENSMUST00000230248.2 ENSMUST00000072776.5 ENSMUST00000229911.2 |
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr4_-_150087587 | 4.96 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr5_+_87148697 | 4.95 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr5_-_87402659 | 4.74 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr16_-_26190578 | 4.68 |
ENSMUST00000023154.3
|
Cldn1
|
claudin 1 |
| chr1_+_172525613 | 4.43 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr13_-_56696222 | 4.38 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr6_+_116627567 | 4.23 |
ENSMUST00000067354.10
ENSMUST00000178241.4 |
Depp1
|
DEPP1 autophagy regulator |
| chr5_-_18054781 | 4.15 |
ENSMUST00000170051.8
|
Cd36
|
CD36 molecule |
| chr5_-_18054702 | 4.15 |
ENSMUST00000165232.8
|
Cd36
|
CD36 molecule |
| chr6_+_42222841 | 3.96 |
ENSMUST00000031897.8
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr9_-_103165423 | 3.72 |
ENSMUST00000123530.8
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr19_-_39637489 | 3.67 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr15_+_4756684 | 3.51 |
ENSMUST00000161997.8
ENSMUST00000022788.15 |
C6
|
complement component 6 |
| chr19_-_39729431 | 3.35 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr16_-_18232202 | 3.20 |
ENSMUST00000165430.8
ENSMUST00000147720.3 |
Comt
|
catechol-O-methyltransferase |
| chrM_+_9870 | 3.03 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chrM_+_10167 | 2.98 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr15_+_4756657 | 2.96 |
ENSMUST00000162585.8
|
C6
|
complement component 6 |
| chr8_+_46984016 | 2.93 |
ENSMUST00000152423.2
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr4_+_137589548 | 2.86 |
ENSMUST00000102518.10
|
Ece1
|
endothelin converting enzyme 1 |
| chr13_+_4283729 | 2.74 |
ENSMUST00000081326.7
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr13_-_32967937 | 2.72 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr6_+_116627635 | 2.52 |
ENSMUST00000204555.2
|
Depp1
|
DEPP1 autophagy regulator |
| chr4_+_102971909 | 2.26 |
ENSMUST00000143417.8
|
Mier1
|
MEIR1 treanscription regulator |
| chrM_+_9459 | 2.25 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr15_+_78290975 | 2.22 |
ENSMUST00000043865.8
ENSMUST00000231159.2 ENSMUST00000169133.8 |
Mpst
|
mercaptopyruvate sulfurtransferase |
| chrX_-_163763337 | 2.22 |
ENSMUST00000112248.9
|
Mospd2
|
motile sperm domain containing 2 |
| chr1_-_173703424 | 2.21 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr2_-_52225146 | 2.21 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr15_+_78290896 | 2.20 |
ENSMUST00000167140.8
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr16_+_96162854 | 2.02 |
ENSMUST00000113794.8
|
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr7_-_19410749 | 2.01 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
| chr3_+_75655504 | 2.00 |
ENSMUST00000189155.4
|
Gm29133
|
predicted gene 29133 |
| chr8_+_61940749 | 1.97 |
ENSMUST00000034058.14
ENSMUST00000126575.2 |
Cbr4
|
carbonyl reductase 4 |
| chr14_-_54949596 | 1.96 |
ENSMUST00000064290.8
|
Cebpe
|
CCAAT/enhancer binding protein (C/EBP), epsilon |
| chr17_+_47083561 | 1.93 |
ENSMUST00000071430.7
|
2310039H08Rik
|
RIKEN cDNA 2310039H08 gene |
| chr6_+_41498716 | 1.85 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr13_+_74554509 | 1.84 |
ENSMUST00000222435.2
|
Ftl1-ps1
|
ferritin light polypeptide 1, pseudogene 1 |
| chr1_+_178146689 | 1.82 |
ENSMUST00000027781.7
|
Cox20
|
cytochrome c oxidase assembly protein 20 |
| chr2_+_87574098 | 1.71 |
ENSMUST00000214723.2
|
Olfr1140
|
olfactory receptor 1140 |
| chr5_-_151510389 | 1.66 |
ENSMUST00000165928.4
|
Vmn2r18
|
vomeronasal 2, receptor 18 |
| chr7_-_14226851 | 1.63 |
ENSMUST00000108524.4
ENSMUST00000211740.2 ENSMUST00000209744.2 |
Sult2a7
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 7 |
| chr6_+_40619913 | 1.62 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chrX_-_16683578 | 1.61 |
ENSMUST00000040820.13
|
Maob
|
monoamine oxidase B |
| chr3_-_63391300 | 1.61 |
ENSMUST00000192926.2
|
Strit1
|
small transmembrane regulator of ion transport 1 |
| chr5_+_36622342 | 1.61 |
ENSMUST00000031099.4
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr3_+_19241484 | 1.59 |
ENSMUST00000130806.8
ENSMUST00000117529.8 ENSMUST00000119865.8 |
Mtfr1
|
mitochondrial fission regulator 1 |
| chr12_+_84332006 | 1.58 |
ENSMUST00000123614.8
ENSMUST00000147363.8 ENSMUST00000135001.8 ENSMUST00000146377.8 |
Ptgr2
|
prostaglandin reductase 2 |
| chr1_-_173707677 | 1.57 |
ENSMUST00000190651.4
ENSMUST00000188804.7 |
Mndal
|
myeloid nuclear differentiation antigen like |
| chr8_-_76133212 | 1.56 |
ENSMUST00000212864.2
|
Gm10358
|
predicted gene 10358 |
| chr7_-_30325514 | 1.55 |
ENSMUST00000208838.2
|
Cox6b1
|
cytochrome c oxidase, subunit 6B1 |
| chr2_-_93988229 | 1.55 |
ENSMUST00000028619.5
|
Hsd17b12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr6_+_41258194 | 1.53 |
ENSMUST00000191646.6
ENSMUST00000103282.3 |
Trbv30
|
T cell receptor beta, variable 30 |
| chr16_-_58860130 | 1.50 |
ENSMUST00000207673.4
|
Olfr187
|
olfactory receptor 187 |
| chr7_-_101518217 | 1.44 |
ENSMUST00000123321.8
|
Folr1
|
folate receptor 1 (adult) |
| chr7_-_101517874 | 1.44 |
ENSMUST00000150184.2
|
Folr1
|
folate receptor 1 (adult) |
| chr3_+_135191366 | 1.43 |
ENSMUST00000029814.10
|
Manba
|
mannosidase, beta A, lysosomal |
| chr3_+_85481416 | 1.42 |
ENSMUST00000107672.8
ENSMUST00000127348.8 ENSMUST00000107674.2 |
Gatb
|
glutamyl-tRNA(Gln) amidotransferase, subunit B |
| chr11_+_81926394 | 1.41 |
ENSMUST00000000193.6
|
Ccl2
|
chemokine (C-C motif) ligand 2 |
| chr7_+_27207226 | 1.38 |
ENSMUST00000125990.2
ENSMUST00000065487.7 |
Prx
|
periaxin |
| chr9_+_107445101 | 1.37 |
ENSMUST00000192887.6
ENSMUST00000195752.6 |
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr19_+_37674029 | 1.36 |
ENSMUST00000073391.5
|
Cyp26c1
|
cytochrome P450, family 26, subfamily c, polypeptide 1 |
| chr4_-_52911794 | 1.36 |
ENSMUST00000051600.3
|
Olfr272
|
olfactory receptor 272 |
| chr11_-_83193412 | 1.33 |
ENSMUST00000176374.2
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr7_-_84328553 | 1.31 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr6_+_41369290 | 1.30 |
ENSMUST00000049079.9
|
Gm5771
|
predicted gene 5771 |
| chr12_+_84147571 | 1.29 |
ENSMUST00000222921.2
|
Acot6
|
acyl-CoA thioesterase 6 |
| chrX_+_41149264 | 1.25 |
ENSMUST00000224454.2
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr7_-_102376698 | 1.25 |
ENSMUST00000215657.2
|
Olfr559
|
olfactory receptor 559 |
| chr7_+_3620356 | 1.24 |
ENSMUST00000076657.11
ENSMUST00000108644.8 |
Ndufa3
|
NADH:ubiquinone oxidoreductase subunit A3 |
| chr1_+_87983099 | 1.23 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr17_+_28075415 | 1.18 |
ENSMUST00000114849.3
|
Uhrf1bp1
|
UHRF1 (ICBP90) binding protein 1 |
| chr1_+_40305738 | 1.16 |
ENSMUST00000114795.3
|
Il1r1
|
interleukin 1 receptor, type I |
| chr15_-_96597610 | 1.14 |
ENSMUST00000023099.8
|
Slc38a2
|
solute carrier family 38, member 2 |
| chr7_-_104250951 | 1.12 |
ENSMUST00000216750.2
ENSMUST00000215538.2 |
Olfr655
|
olfactory receptor 655 |
| chr16_+_57173632 | 1.11 |
ENSMUST00000099667.3
|
Filip1l
|
filamin A interacting protein 1-like |
| chr2_+_67935015 | 1.11 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr13_+_6598185 | 1.10 |
ENSMUST00000021611.10
ENSMUST00000222485.2 |
Pitrm1
|
pitrilysin metallepetidase 1 |
| chrX_-_166047275 | 1.09 |
ENSMUST00000112170.2
|
Tlr8
|
toll-like receptor 8 |
| chr9_+_37957851 | 1.07 |
ENSMUST00000181088.4
|
Olfr884
|
olfactory receptor 884 |
| chr10_-_129630735 | 1.05 |
ENSMUST00000217283.2
ENSMUST00000214206.2 ENSMUST00000214878.2 |
Olfr810
|
olfactory receptor 810 |
| chr6_+_41435846 | 1.05 |
ENSMUST00000031910.8
|
Prss1
|
protease, serine 1 (trypsin 1) |
| chr7_-_10638954 | 1.05 |
ENSMUST00000210847.2
ENSMUST00000168158.3 |
Zscan4b
|
zinc finger and SCAN domain containing 4B |
| chr3_+_40801405 | 1.05 |
ENSMUST00000108078.9
|
Abhd18
|
abhydrolase domain containing 18 |
| chr7_-_9575286 | 1.04 |
ENSMUST00000174433.2
|
Gm10302
|
predicted gene 10302 |
| chr14_-_30723549 | 1.04 |
ENSMUST00000226782.2
ENSMUST00000186131.7 ENSMUST00000228767.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chrX_-_36854522 | 1.02 |
ENSMUST00000184088.2
|
Rhox2h
|
reproductive homeobox 2H |
| chr4_-_113144152 | 1.02 |
ENSMUST00000138966.9
|
Skint6
|
selection and upkeep of intraepithelial T cells 6 |
| chr7_-_11044586 | 1.01 |
ENSMUST00000166753.4
ENSMUST00000210521.2 |
Zscan4e
|
zinc finger and SCAN domain containing 4E |
| chr4_-_144513121 | 1.00 |
ENSMUST00000105746.3
|
Aadacl4fm5
|
AADACL4 family member 5 |
| chr7_-_10900086 | 1.00 |
ENSMUST00000067210.12
|
Zscan4d
|
zinc finger and SCAN domain containing 4D |
| chr6_+_35229589 | 0.98 |
ENSMUST00000152147.8
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr16_+_29398165 | 0.95 |
ENSMUST00000161186.8
ENSMUST00000038867.13 |
Opa1
|
OPA1, mitochondrial dynamin like GTPase |
| chr13_+_28033322 | 0.95 |
ENSMUST00000110335.10
ENSMUST00000054932.13 |
Prl2c1
|
Prolactin family 2, subfamily c, member 1 |
| chr14_+_55842002 | 0.94 |
ENSMUST00000138037.2
|
Irf9
|
interferon regulatory factor 9 |
| chr7_+_23451695 | 0.93 |
ENSMUST00000228331.2
|
Vmn1r174
|
vomeronasal 1 receptor 174 |
| chr7_-_8492074 | 0.92 |
ENSMUST00000164845.4
|
Vmn2r45
|
vomeronasal 2, receptor 45 |
| chr11_-_99519057 | 0.91 |
ENSMUST00000081007.7
|
Krtap4-1
|
keratin associated protein 4-1 |
| chr7_+_11343235 | 0.91 |
ENSMUST00000210559.2
|
Zscan4-ps3
|
zinc finger and SCAN domain containing 4, pseudogene 3 |
| chr7_+_86169176 | 0.90 |
ENSMUST00000172965.3
|
Olfr297
|
olfactory receptor 297 |
| chr7_-_8203319 | 0.90 |
ENSMUST00000086282.13
ENSMUST00000146278.9 ENSMUST00000142934.3 |
Vmn2r42
|
vomeronasal 2, receptor 42 |
| chr2_-_36975758 | 0.89 |
ENSMUST00000100145.2
|
Olfr361
|
olfactory receptor 361 |
| chr14_+_53607470 | 0.88 |
ENSMUST00000103652.5
|
Trav14n-3
|
T cell receptor alpha variable 14N-3 |
| chr1_+_87983189 | 0.88 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr17_+_73414977 | 0.88 |
ENSMUST00000130574.4
ENSMUST00000149064.9 ENSMUST00000067545.8 |
Lclat1
|
lysocardiolipin acyltransferase 1 |
| chr17_+_38143840 | 0.88 |
ENSMUST00000213857.2
|
Olfr125
|
olfactory receptor 125 |
| chr6_-_70021662 | 0.87 |
ENSMUST00000196959.2
|
Igkv8-34
|
immunoglobulin kappa variable 8-34 |
| chr17_-_37594351 | 0.86 |
ENSMUST00000216328.2
|
Olfr99
|
olfactory receptor 99 |
| chr7_+_104963189 | 0.85 |
ENSMUST00000098153.3
|
Olfr689
|
olfactory receptor 689 |
| chr14_+_53743184 | 0.85 |
ENSMUST00000103583.5
|
Trav10
|
T cell receptor alpha variable 10 |
| chr12_+_8258107 | 0.85 |
ENSMUST00000037383.13
ENSMUST00000218883.2 ENSMUST00000218086.2 ENSMUST00000169104.3 ENSMUST00000217999.2 |
Ldah
|
lipid droplet associated hydrolase |
| chr4_+_114020581 | 0.85 |
ENSMUST00000079915.10
ENSMUST00000164297.8 |
Skint11
|
selection and upkeep of intraepithelial T cells 11 |
| chr7_-_9226652 | 0.85 |
ENSMUST00000072787.5
|
Vmn2r37
|
vomeronasal 2, receptor 37 |
| chr7_+_21006104 | 0.84 |
ENSMUST00000098739.3
|
Vmn1r125
|
vomeronasal 1 receptor 125 |
| chr14_-_54520382 | 0.84 |
ENSMUST00000059996.7
|
Olfr49
|
olfactory receptor 49 |
| chr11_+_100225233 | 0.84 |
ENSMUST00000017309.2
|
Gast
|
gastrin |
| chr1_+_163889713 | 0.83 |
ENSMUST00000097491.10
|
Sell
|
selectin, lymphocyte |
| chr2_+_26800757 | 0.83 |
ENSMUST00000102898.5
|
Rpl7a
|
ribosomal protein L7A |
| chr16_-_18407558 | 0.82 |
ENSMUST00000232589.2
|
Tbx1
|
T-box 1 |
| chr4_+_130001349 | 0.82 |
ENSMUST00000030563.6
|
Pef1
|
penta-EF hand domain containing 1 |
| chr7_+_11131842 | 0.81 |
ENSMUST00000145237.8
ENSMUST00000141491.2 |
Zscan4f
|
zinc finger and SCAN domain containing 4F |
| chr9_+_3013140 | 0.81 |
ENSMUST00000143083.3
|
Gm10721
|
predicted gene 10721 |
| chr1_+_74627506 | 0.81 |
ENSMUST00000113732.2
|
Bcs1l
|
BCS1-like (yeast) |
| chr2_-_37007795 | 0.81 |
ENSMUST00000213817.2
ENSMUST00000215927.2 |
Olfr362
|
olfactory receptor 362 |
| chr15_-_57128522 | 0.80 |
ENSMUST00000137764.2
ENSMUST00000022995.13 |
Slc22a22
|
solute carrier family 22 (organic cation transporter), member 22 |
| chr14_+_53655806 | 0.80 |
ENSMUST00000103637.6
|
Trav4-2
|
T cell receptor alpha variable 4-2 |
| chr3_+_53752507 | 0.79 |
ENSMUST00000108014.2
|
Gm10985
|
predicted gene 10985 |
| chr1_-_155108455 | 0.79 |
ENSMUST00000035914.5
|
BC034090
|
cDNA sequence BC034090 |
| chr2_-_90087936 | 0.78 |
ENSMUST00000213968.2
|
Olfr142
|
olfactory receptor 142 |
| chr10_-_129948657 | 0.78 |
ENSMUST00000081469.2
|
Olfr823
|
olfactory receptor 823 |
| chr3_+_40848580 | 0.77 |
ENSMUST00000159774.7
ENSMUST00000203472.3 ENSMUST00000203650.3 ENSMUST00000108077.10 ENSMUST00000203892.2 |
Abhd18
|
abhydrolase domain containing 18 |
| chr6_-_40976413 | 0.77 |
ENSMUST00000166306.3
|
Gm2663
|
predicted gene 2663 |
| chr17_+_28075495 | 0.76 |
ENSMUST00000233809.2
|
Uhrf1bp1
|
UHRF1 (ICBP90) binding protein 1 |
| chr4_+_111830119 | 0.75 |
ENSMUST00000106568.8
ENSMUST00000055014.11 ENSMUST00000163281.2 |
Skint7
|
selection and upkeep of intraepithelial T cells 7 |
| chr7_+_12034357 | 0.75 |
ENSMUST00000072801.5
ENSMUST00000227672.2 |
Vmn1r82
|
vomeronasal 1 receptor 82 |
| chr7_-_7340608 | 0.75 |
ENSMUST00000174368.9
ENSMUST00000072475.9 |
Vmn2r30
|
vomeronasal 2, receptor 30 |
| chr12_+_8258166 | 0.75 |
ENSMUST00000220274.2
|
Ldah
|
lipid droplet associated hydrolase |
| chr11_+_58062467 | 0.74 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr7_-_7692512 | 0.73 |
ENSMUST00000173459.3
|
Vmn2r34
|
vomeronasal 2, receptor 34 |
| chr6_-_41012435 | 0.71 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr13_+_21938258 | 0.71 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15 |
| chr7_+_127368981 | 0.71 |
ENSMUST00000118865.8
ENSMUST00000061587.13 ENSMUST00000121504.2 |
Orai3
|
ORAI calcium release-activated calcium modulator 3 |
| chr7_+_11248212 | 0.71 |
ENSMUST00000091440.4
|
Zscan4-ps2
|
zinc finger and SCAN domain containing 4, pseudogene 2 |
| chr15_+_80832685 | 0.70 |
ENSMUST00000023043.10
ENSMUST00000164806.6 ENSMUST00000207170.2 ENSMUST00000168756.8 |
Adsl
|
adenylosuccinate lyase |
| chr2_-_88942854 | 0.70 |
ENSMUST00000099795.2
|
Olfr1221
|
olfactory receptor 1221 |
| chr6_+_123679629 | 0.68 |
ENSMUST00000172391.4
|
Vmn2r23
|
vomeronasal 2, receptor 23 |
| chr2_-_87543523 | 0.68 |
ENSMUST00000214209.2
|
Olfr1137
|
olfactory receptor 1137 |
| chr6_-_69477770 | 0.67 |
ENSMUST00000197448.2
|
Igkv4-58
|
immunoglobulin kappa variable 4-58 |
| chr7_+_41909477 | 0.66 |
ENSMUST00000166131.2
|
Vmn2r61
|
vomeronasal 2, receptor 61 |
| chr7_-_103420801 | 0.66 |
ENSMUST00000106878.3
|
Olfr69
|
olfactory receptor 69 |
| chr8_-_3928495 | 0.66 |
ENSMUST00000209176.2
ENSMUST00000011445.8 |
Cd209d
|
CD209d antigen |
| chr9_+_3025417 | 0.66 |
ENSMUST00000075573.7
|
Gm10717
|
predicted gene 10717 |
| chr3_-_16060545 | 0.66 |
ENSMUST00000194367.6
|
Gm5150
|
predicted gene 5150 |
| chr11_+_73960781 | 0.65 |
ENSMUST00000108463.8
ENSMUST00000074813.12 |
Zfp616
|
zinc finger protein 616 |
| chr2_-_18053595 | 0.65 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr10_+_125802084 | 0.65 |
ENSMUST00000074807.8
|
Lrig3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr9_+_3018753 | 0.64 |
ENSMUST00000179272.2
|
Gm10719
|
predicted gene 10719 |
| chr11_+_73961011 | 0.64 |
ENSMUST00000116546.3
|
Zfp616
|
zinc finger protein 616 |
| chr14_-_42463874 | 0.64 |
ENSMUST00000100697.4
|
Gm3633
|
predicted gene 3633 |
| chrX_+_138963623 | 0.64 |
ENSMUST00000044806.3
|
Dnaaf6b
|
dynein axonemal assembly factor 6B |
| chr19_+_13745671 | 0.64 |
ENSMUST00000061669.3
|
Olfr1495
|
olfactory receptor 1495 |
| chr7_+_143376871 | 0.63 |
ENSMUST00000128454.8
ENSMUST00000073878.12 |
Dhcr7
|
7-dehydrocholesterol reductase |
| chr17_+_26342474 | 0.63 |
ENSMUST00000025014.10
ENSMUST00000236166.2 ENSMUST00000127647.3 |
Mrpl28
|
mitochondrial ribosomal protein L28 |
| chr7_+_23331413 | 0.63 |
ENSMUST00000078458.4
|
Vmn1r171
|
vomeronasal 1 receptor 171 |
| chr7_-_42442949 | 0.62 |
ENSMUST00000169130.3
|
Vmn2r62
|
vomeronasal 2, receptor 62 |
| chr7_-_104677667 | 0.61 |
ENSMUST00000215899.2
ENSMUST00000214318.3 |
Olfr675
|
olfactory receptor 675 |
| chr7_-_9839668 | 0.61 |
ENSMUST00000094863.6
|
Vmn2r51
|
vomeronasal 2, receptor 51 |
| chr7_+_10739672 | 0.61 |
ENSMUST00000131379.4
|
Zscan4c
|
zinc finger and SCAN domain containing 4C |
| chr14_-_52616625 | 0.61 |
ENSMUST00000214980.2
|
Olfr1512
|
olfactory receptor 1512 |
| chr14_-_62693735 | 0.59 |
ENSMUST00000165651.8
ENSMUST00000022501.10 |
Gucy1b2
|
guanylate cyclase 1, soluble, beta 2 |
| chr4_+_52964547 | 0.59 |
ENSMUST00000215010.2
ENSMUST00000215127.2 |
Olfr270
|
olfactory receptor 270 |
| chr6_-_128558560 | 0.57 |
ENSMUST00000060574.9
|
A2ml1
|
alpha-2-macroglobulin like 1 |
| chr7_+_6233178 | 0.57 |
ENSMUST00000165445.3
|
Zscan5b
|
zinc finger and SCAN domain containing 5B |
| chr4_+_57637817 | 0.56 |
ENSMUST00000150412.4
|
Pakap
|
paralemmin A kinase anchor protein |
| chr3_+_144283355 | 0.56 |
ENSMUST00000151086.3
|
Selenof
|
selenoprotein F |
| chr7_+_20505110 | 0.56 |
ENSMUST00000174538.2
|
Vmn1r112
|
vomeronasal 1 receptor 112 |
| chr7_-_7822866 | 0.55 |
ENSMUST00000169683.2
|
Vmn2r35
|
vomeronasal 2, receptor 35 |
| chr11_-_22932090 | 0.54 |
ENSMUST00000160826.2
ENSMUST00000093270.6 ENSMUST00000071068.9 ENSMUST00000159081.8 |
Commd1b
Commd1
|
COMM domain containing 1B COMM domain containing 1 |
| chr4_-_58912678 | 0.54 |
ENSMUST00000144512.8
ENSMUST00000102889.10 ENSMUST00000055822.15 |
Ecpas
|
Ecm29 proteasome adaptor and scaffold |
| chr7_-_103191924 | 0.54 |
ENSMUST00000214269.3
|
Olfr612
|
olfactory receptor 612 |
| chr1_+_116730713 | 0.54 |
ENSMUST00000179777.3
|
Gm19965
|
predicted gene, 19965 |
| chr2_-_98497609 | 0.53 |
ENSMUST00000099683.2
|
Gm10800
|
predicted gene 10800 |
| chr9_+_3023547 | 0.53 |
ENSMUST00000099046.4
|
Gm10718
|
predicted gene 10718 |
| chr5_+_35740371 | 0.52 |
ENSMUST00000068947.14
ENSMUST00000114237.8 ENSMUST00000156125.8 ENSMUST00000202266.4 ENSMUST00000068563.12 |
Acox3
|
acyl-Coenzyme A oxidase 3, pristanoyl |
| chr12_-_112831848 | 0.52 |
ENSMUST00000222776.2
|
Gpr132
|
G protein-coupled receptor 132 |
| chr6_+_125016723 | 0.51 |
ENSMUST00000140131.8
ENSMUST00000032480.14 |
Ing4
|
inhibitor of growth family, member 4 |
| chr2_+_85836999 | 0.50 |
ENSMUST00000079298.5
|
Olfr1032
|
olfactory receptor 1032 |
| chr14_-_42119315 | 0.49 |
ENSMUST00000169972.2
|
Gm9611
|
predicted gene 9611 |
| chr7_+_86053054 | 0.49 |
ENSMUST00000174362.3
|
Olfr301
|
olfactory receptor 301 |
| chr2_+_88217406 | 0.49 |
ENSMUST00000214040.3
|
Olfr1178
|
olfactory receptor 1178 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 1.8 | 5.5 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 1.8 | 5.5 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 1.7 | 8.3 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 1.6 | 6.5 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.6 | 4.7 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 1.5 | 7.4 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 1.0 | 2.9 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.7 | 7.9 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.6 | 3.2 | GO:0045963 | regulation of sulfur amino acid metabolic process(GO:0031335) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.6 | 4.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.5 | 2.0 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.5 | 1.4 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.5 | 1.4 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) monocyte homeostasis(GO:0035702) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.4 | 4.4 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.4 | 1.6 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.3 | 1.4 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.3 | 2.9 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.3 | 1.3 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.3 | 1.2 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.3 | 2.0 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.3 | 0.8 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.3 | 0.8 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.3 | 2.9 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.3 | 1.3 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.3 | 1.5 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.2 | 5.0 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.2 | 7.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 1.4 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.2 | 0.7 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.6 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.2 | 0.6 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.2 | 1.4 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 0.5 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.2 | 2.1 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.7 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 0.8 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.2 | 2.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 2.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 5.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.6 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.3 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.1 | 1.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 1.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.3 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 2.6 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.3 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.1 | 0.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 1.4 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 5.9 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.2 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 1.6 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 3.9 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 1.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.2 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.1 | 1.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.7 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 4.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.4 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 2.0 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 1.6 | GO:0090077 | foam cell differentiation(GO:0090077) |
| 0.0 | 1.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 1.2 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.6 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 1.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 1.6 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.3 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.8 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.3 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 2.2 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.8 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:0070537 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.3 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 20.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 1.0 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.5 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 1.3 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.6 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 2.9 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.7 | 6.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 1.6 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.5 | 1.4 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.3 | 1.3 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.3 | 2.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 1.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 2.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 1.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 4.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 6.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 8.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 32.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 0.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 6.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 4.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 2.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 2.0 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 2.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 4.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 1.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0044438 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.1 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 3.0 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 27.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 2.0 | 7.9 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 1.8 | 5.5 | GO:0070540 | stearic acid binding(GO:0070540) |
| 1.7 | 8.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 1.7 | 5.0 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 1.5 | 4.4 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 1.1 | 45.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.1 | 3.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.8 | 7.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.6 | 2.9 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.6 | 4.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.5 | 1.5 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.5 | 1.4 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.4 | 1.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 1.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.3 | 2.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 1.6 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.3 | 2.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 1.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.3 | 6.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.3 | 5.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 1.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.3 | 8.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 1.6 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.3 | 5.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.3 | 2.9 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 1.4 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 7.0 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 0.6 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.2 | 4.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 1.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 0.7 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 4.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 2.0 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 1.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.6 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 1.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 2.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.8 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 1.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.8 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.1 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 1.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.3 | GO:0001179 | RNA polymerase I transcription factor binding(GO:0001179) |
| 0.0 | 0.3 | GO:0001601 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 4.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 5.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.8 | GO:0019840 | isoprenoid binding(GO:0019840) |
| 0.0 | 1.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 19.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 5.0 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 1.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.6 | GO:0016298 | lipase activity(GO:0016298) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 11.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 0.9 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 4.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 4.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 4.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.5 | 5.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 1.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 6.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 5.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 4.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.9 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 4.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 2.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 8.8 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 7.6 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 1.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 13.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 3.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 2.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 3.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 2.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |