Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
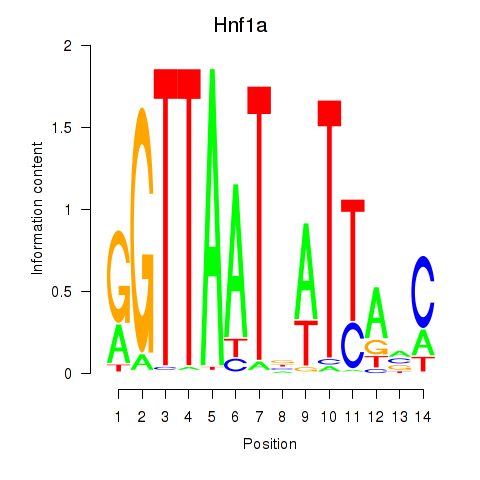
Results for Hnf1a
Z-value: 5.10
Transcription factors associated with Hnf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hnf1a
|
ENSMUSG00000029556.13 | Hnf1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hnf1a | mm39_v1_chr5_-_115109118_115109138 | 0.85 | 7.7e-21 | Click! |
Activity profile of Hnf1a motif
Sorted Z-values of Hnf1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hnf1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_90666791 | 128.24 |
ENSMUST00000113179.9
ENSMUST00000128740.2 |
Afm
|
afamin |
| chr2_+_163389068 | 113.28 |
ENSMUST00000109411.8
ENSMUST00000018094.13 |
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr12_-_103739847 | 99.65 |
ENSMUST00000078869.6
|
Serpina1d
|
serine (or cysteine) peptidase inhibitor, clade A, member 1D |
| chr12_-_103829810 | 96.60 |
ENSMUST00000085056.8
ENSMUST00000072876.12 ENSMUST00000124717.2 |
Serpina1a
|
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
| chr5_-_89605622 | 95.68 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr12_-_103704417 | 90.09 |
ENSMUST00000095450.11
ENSMUST00000187220.2 |
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr19_+_42034231 | 80.96 |
ENSMUST00000172244.8
ENSMUST00000081714.5 |
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr17_-_35351026 | 77.74 |
ENSMUST00000025249.7
|
Apom
|
apolipoprotein M |
| chr5_+_90608751 | 76.90 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr1_+_88139678 | 74.59 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr12_-_103923145 | 73.48 |
ENSMUST00000085054.5
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr17_+_12597490 | 72.10 |
ENSMUST00000014578.7
|
Plg
|
plasminogen |
| chr19_+_56276375 | 72.06 |
ENSMUST00000166049.8
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr11_+_101258368 | 71.94 |
ENSMUST00000019469.3
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr12_-_103871146 | 71.84 |
ENSMUST00000074051.6
|
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1C |
| chr2_+_163348728 | 71.69 |
ENSMUST00000143911.8
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr3_-_82957104 | 67.99 |
ENSMUST00000048246.5
|
Fgb
|
fibrinogen beta chain |
| chr2_-_91466739 | 64.17 |
ENSMUST00000111335.2
ENSMUST00000028681.15 |
F2
|
coagulation factor II |
| chr10_+_60925130 | 63.72 |
ENSMUST00000020298.8
|
Pcbd1
|
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 1 |
| chr5_+_35198853 | 62.44 |
ENSMUST00000030985.10
ENSMUST00000202573.2 |
Hgfac
|
hepatocyte growth factor activator |
| chr6_+_141575226 | 62.07 |
ENSMUST00000042812.9
|
Slco1b2
|
solute carrier organic anion transporter family, member 1b2 |
| chr19_+_56276343 | 61.68 |
ENSMUST00000095948.11
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr3_+_129630380 | 59.70 |
ENSMUST00000077918.7
|
Cfi
|
complement component factor i |
| chr12_-_103623418 | 59.09 |
ENSMUST00000044159.7
|
Serpina6
|
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
| chr10_+_60925108 | 58.01 |
ENSMUST00000218005.2
|
Pcbd1
|
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 1 |
| chr7_+_30193047 | 51.87 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr2_+_162829250 | 51.39 |
ENSMUST00000018012.14
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chrX_+_100420873 | 51.34 |
ENSMUST00000052130.14
|
Gjb1
|
gap junction protein, beta 1 |
| chr1_+_172525613 | 51.06 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr1_-_130589349 | 48.64 |
ENSMUST00000027657.14
|
C4bp
|
complement component 4 binding protein |
| chr7_+_140343652 | 48.38 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr2_-_34990689 | 46.94 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr1_-_130589321 | 46.26 |
ENSMUST00000137276.3
|
C4bp
|
complement component 4 binding protein |
| chr9_+_21746785 | 45.55 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr3_+_82933383 | 44.92 |
ENSMUST00000029630.15
ENSMUST00000166581.4 |
Fga
|
fibrinogen alpha chain |
| chr11_-_11848107 | 44.73 |
ENSMUST00000178704.8
|
Ddc
|
dopa decarboxylase |
| chr2_+_162829422 | 44.22 |
ENSMUST00000117123.2
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr12_-_103597663 | 43.34 |
ENSMUST00000121625.2
ENSMUST00000044231.12 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr1_+_133237516 | 40.73 |
ENSMUST00000094557.7
ENSMUST00000192465.2 ENSMUST00000193888.6 ENSMUST00000194044.6 ENSMUST00000184603.8 |
Golt1a
Gm28040
Gm28040
|
golgi transport 1A predicted gene, 28040 predicted gene, 28040 |
| chr4_+_138694422 | 38.67 |
ENSMUST00000116094.5
ENSMUST00000239443.2 |
Rnf186
|
ring finger protein 186 |
| chr5_+_33261563 | 38.60 |
ENSMUST00000011178.5
|
Slc5a1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr14_+_51333816 | 38.53 |
ENSMUST00000169895.3
|
Rnase4
|
ribonuclease, RNase A family 4 |
| chr11_-_11848044 | 38.14 |
ENSMUST00000066237.10
|
Ddc
|
dopa decarboxylase |
| chr1_+_88066086 | 37.68 |
ENSMUST00000014263.6
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr7_-_79492091 | 37.13 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr2_-_34951443 | 36.57 |
ENSMUST00000028233.7
|
Hc
|
hemolytic complement |
| chr15_+_10216041 | 35.90 |
ENSMUST00000130720.8
|
Prlr
|
prolactin receptor |
| chr9_-_70841881 | 35.17 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chrX_+_100419965 | 34.35 |
ENSMUST00000119080.8
|
Gjb1
|
gap junction protein, beta 1 |
| chr1_+_87998487 | 33.62 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr9_-_106035308 | 32.95 |
ENSMUST00000159809.2
ENSMUST00000162562.2 ENSMUST00000036382.13 |
Glyctk
|
glycerate kinase |
| chr9_-_70842090 | 32.08 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr5_+_31454787 | 31.23 |
ENSMUST00000201166.4
ENSMUST00000072228.9 |
Gckr
|
glucokinase regulatory protein |
| chr11_+_70410445 | 29.54 |
ENSMUST00000179000.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr8_-_122671588 | 29.40 |
ENSMUST00000057653.8
|
Car5a
|
carbonic anhydrase 5a, mitochondrial |
| chr13_+_55300453 | 29.39 |
ENSMUST00000005452.6
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr13_+_54849268 | 29.16 |
ENSMUST00000037145.8
|
Cdhr2
|
cadherin-related family member 2 |
| chr4_+_150938376 | 29.03 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr5_+_31454939 | 28.98 |
ENSMUST00000201675.3
|
Gckr
|
glucokinase regulatory protein |
| chr11_+_7147779 | 28.89 |
ENSMUST00000020704.8
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr9_-_106035332 | 28.16 |
ENSMUST00000112543.9
|
Glyctk
|
glycerate kinase |
| chr16_-_38253507 | 27.75 |
ENSMUST00000002926.8
|
Pla1a
|
phospholipase A1 member A |
| chr10_+_34359513 | 27.01 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chr10_+_34359395 | 25.81 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr7_+_127865472 | 25.80 |
ENSMUST00000033045.11
|
Slc5a2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr3_+_3573084 | 25.08 |
ENSMUST00000108393.8
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr7_+_140795866 | 24.97 |
ENSMUST00000210993.2
ENSMUST00000133763.8 |
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr11_-_78313043 | 24.24 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr7_+_51537645 | 24.07 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr1_-_173105344 | 23.67 |
ENSMUST00000111224.5
|
Mptx2
|
mucosal pentraxin 2 |
| chr7_+_140796537 | 23.41 |
ENSMUST00000141804.2
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr11_-_84058292 | 23.29 |
ENSMUST00000050771.8
|
Gm11437
|
predicted gene 11437 |
| chr18_-_43870622 | 21.67 |
ENSMUST00000025381.4
|
Spink1
|
serine peptidase inhibitor, Kazal type 1 |
| chr4_-_94538370 | 21.60 |
ENSMUST00000053419.9
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr2_+_110427643 | 21.38 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr7_+_140796096 | 21.28 |
ENSMUST00000153081.2
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr7_-_119078472 | 21.14 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr9_-_76953070 | 20.99 |
ENSMUST00000034911.7
|
Tinag
|
tubulointerstitial nephritis antigen |
| chr17_+_12803019 | 20.09 |
ENSMUST00000046959.9
ENSMUST00000233066.2 |
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chrX_+_162873183 | 19.96 |
ENSMUST00000015545.10
|
Cltrn
|
collectrin, amino acid transport regulator |
| chr19_-_43879031 | 19.38 |
ENSMUST00000212048.2
|
Dnmbp
|
dynamin binding protein |
| chr1_-_20688196 | 19.14 |
ENSMUST00000088448.12
|
Pkhd1
|
polycystic kidney and hepatic disease 1 |
| chr10_+_87695117 | 18.73 |
ENSMUST00000105300.9
|
Igf1
|
insulin-like growth factor 1 |
| chr7_-_119078330 | 18.28 |
ENSMUST00000207460.2
|
Umod
|
uromodulin |
| chr15_-_58261093 | 17.80 |
ENSMUST00000227274.3
|
Anxa13
|
annexin A13 |
| chr10_+_87694924 | 17.78 |
ENSMUST00000095360.11
|
Igf1
|
insulin-like growth factor 1 |
| chr12_-_103623354 | 16.66 |
ENSMUST00000152517.2
|
Serpina6
|
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
| chr11_+_83742961 | 16.60 |
ENSMUST00000146786.8
|
Hnf1b
|
HNF1 homeobox B |
| chr11_+_70410009 | 16.46 |
ENSMUST00000057685.3
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr12_+_59142439 | 15.76 |
ENSMUST00000219140.3
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr2_-_32314017 | 15.72 |
ENSMUST00000113307.9
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr4_-_63090355 | 15.00 |
ENSMUST00000156618.9
ENSMUST00000030042.3 |
Kif12
|
kinesin family member 12 |
| chrX_-_137985960 | 14.81 |
ENSMUST00000033626.15
ENSMUST00000060824.4 |
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr9_-_71070506 | 14.54 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr2_-_134396268 | 13.73 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr6_-_52217821 | 12.55 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr4_+_119494901 | 12.08 |
ENSMUST00000024015.3
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr3_+_14545751 | 11.67 |
ENSMUST00000037321.8
ENSMUST00000120484.8 ENSMUST00000120801.2 |
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr8_-_65489791 | 11.17 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr11_+_115802828 | 10.58 |
ENSMUST00000132961.2
|
Smim6
|
small integral membrane protein 6 |
| chr8_-_105350881 | 8.20 |
ENSMUST00000211903.2
|
Cdh16
|
cadherin 16 |
| chr14_+_80237691 | 7.95 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr8_-_105350533 | 7.35 |
ENSMUST00000212662.2
|
Cdh16
|
cadherin 16 |
| chr8_-_65489834 | 7.17 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr8_-_105350816 | 7.15 |
ENSMUST00000212447.2
|
Cdh16
|
cadherin 16 |
| chrX_-_137985979 | 7.07 |
ENSMUST00000152457.2
|
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr8_-_105350898 | 6.98 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr8_+_117822593 | 6.96 |
ENSMUST00000034308.16
ENSMUST00000176860.2 |
Bco1
|
beta-carotene oxygenase 1 |
| chr8_-_95869114 | 6.74 |
ENSMUST00000212554.2
ENSMUST00000169748.9 ENSMUST00000034240.15 |
Kifc3
|
kinesin family member C3 |
| chr7_-_44711771 | 6.67 |
ENSMUST00000210101.2
ENSMUST00000209219.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr10_-_125164399 | 6.60 |
ENSMUST00000063318.10
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr2_+_74557418 | 6.35 |
ENSMUST00000111980.4
|
Hoxd4
|
homeobox D4 |
| chr8_+_123920682 | 6.12 |
ENSMUST00000212409.2
|
Dpep1
|
dipeptidase 1 |
| chr8_-_105350842 | 5.86 |
ENSMUST00000212324.2
|
Cdh16
|
cadherin 16 |
| chr12_+_76580386 | 5.41 |
ENSMUST00000219063.2
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr8_-_107792264 | 5.20 |
ENSMUST00000034393.7
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr1_+_174158083 | 4.63 |
ENSMUST00000027816.5
|
Mptx1
|
mucosal pentraxin 1 |
| chr7_-_101519872 | 4.51 |
ENSMUST00000106986.9
|
Folr1
|
folate receptor 1 (adult) |
| chr10_+_75983285 | 4.03 |
ENSMUST00000020450.4
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr7_-_101519791 | 3.73 |
ENSMUST00000124026.8
|
Folr1
|
folate receptor 1 (adult) |
| chr5_+_88032867 | 3.54 |
ENSMUST00000129757.9
|
Odam
|
odontogenic, ameloblast asssociated |
| chr8_+_123920233 | 3.45 |
ENSMUST00000212773.2
|
Dpep1
|
dipeptidase 1 |
| chr17_+_71511642 | 3.41 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr11_+_78068931 | 2.79 |
ENSMUST00000147819.8
|
Tlcd1
|
TLC domain containing 1 |
| chr5_+_88635834 | 2.61 |
ENSMUST00000199104.5
ENSMUST00000031222.9 |
Enam
|
enamelin |
| chr10_+_115653152 | 2.51 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr5_-_114796425 | 2.45 |
ENSMUST00000112225.8
ENSMUST00000071968.9 |
Trpv4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chr6_+_29859685 | 2.35 |
ENSMUST00000134438.2
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr6_-_23248361 | 2.08 |
ENSMUST00000031709.7
|
Fezf1
|
Fez family zinc finger 1 |
| chr4_+_11758147 | 1.87 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr7_-_101519914 | 1.84 |
ENSMUST00000106985.8
ENSMUST00000151706.8 |
Folr1
|
folate receptor 1 (adult) |
| chr6_+_29859660 | 1.71 |
ENSMUST00000128927.9
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr11_+_118319029 | 1.05 |
ENSMUST00000124861.2
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr6_+_29859372 | 0.74 |
ENSMUST00000115238.10
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr7_-_103218350 | 0.63 |
ENSMUST00000217293.2
|
Olfr616
|
olfactory receptor 616 |
| chr5_+_122529941 | 0.54 |
ENSMUST00000102525.11
|
Arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr15_+_7840327 | 0.53 |
ENSMUST00000022744.5
|
Gdnf
|
glial cell line derived neurotrophic factor |
| chr18_+_65158873 | 0.46 |
ENSMUST00000226058.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr1_-_82746169 | 0.32 |
ENSMUST00000027331.3
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr19_-_47907628 | 0.27 |
ENSMUST00000237029.2
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 61.7 | 185.0 | GO:0042977 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 28.2 | 112.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 25.9 | 77.7 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 25.6 | 76.9 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 24.9 | 74.6 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 20.2 | 81.0 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 17.3 | 51.9 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 14.4 | 72.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 13.4 | 67.2 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 12.8 | 64.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 12.2 | 121.7 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 11.8 | 82.9 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 9.9 | 39.4 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 9.8 | 29.4 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 8.6 | 60.2 | GO:0009750 | response to fructose(GO:0009750) |
| 7.7 | 38.6 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 7.6 | 45.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 7.3 | 36.6 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 7.3 | 21.9 | GO:0033189 | response to vitamin A(GO:0033189) |
| 7.3 | 29.0 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 7.2 | 71.9 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 7.1 | 85.7 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 6.5 | 71.3 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 6.4 | 51.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 6.0 | 35.9 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 5.8 | 29.2 | GO:1904970 | brush border assembly(GO:1904970) |
| 5.4 | 21.7 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 5.2 | 36.5 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 4.8 | 95.7 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 4.2 | 16.6 | GO:0061206 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 4.0 | 63.9 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 3.6 | 14.5 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 2.7 | 13.7 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 2.7 | 138.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 2.4 | 9.6 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 2.4 | 75.8 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 1.9 | 48.4 | GO:0017144 | drug metabolic process(GO:0017144) |
| 1.8 | 37.1 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 1.5 | 90.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 1.2 | 28.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 1.1 | 20.1 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 1.0 | 35.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 1.0 | 154.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 1.0 | 6.7 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.9 | 6.6 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.9 | 2.6 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.8 | 2.5 | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.8 | 341.6 | GO:0043434 | response to peptide hormone(GO:0043434) |
| 0.7 | 7.9 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.7 | 34.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.6 | 52.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.5 | 15.8 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.4 | 15.7 | GO:0032094 | response to food(GO:0032094) |
| 0.4 | 73.4 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.4 | 7.0 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.4 | 19.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.3 | 12.6 | GO:0060065 | uterus development(GO:0060065) |
| 0.3 | 2.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 21.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 65.9 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.2 | 38.5 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.2 | 45.6 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.2 | 0.5 | GO:0072107 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.1 | 24.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 3.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 34.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 7.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.0 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 2.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 15.0 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 17.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 24.6 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.1 | 134.7 | GO:0006508 | proteolysis(GO:0006508) |
| 0.1 | 8.6 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.1 | 10.7 | GO:0007369 | gastrulation(GO:0007369) |
| 0.1 | 3.5 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 15.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 4.3 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.5 | 77.7 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 12.5 | 112.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 12.0 | 72.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 5.0 | 74.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 4.6 | 36.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 3.7 | 36.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 3.2 | 85.7 | GO:0005922 | connexon complex(GO:0005922) |
| 2.9 | 29.2 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 2.7 | 37.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 1.9 | 67.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 1.4 | 365.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.8 | 27.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.5 | 71.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.5 | 62.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.5 | 107.5 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.5 | 15.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.4 | 6.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.4 | 813.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.4 | 81.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.4 | 239.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.3 | 82.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 29.4 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.2 | 57.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 15.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 105.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 15.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 13.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 223.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 336.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 32.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 37.7 | GO:0005815 | microtubule organizing center(GO:0005815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 61.7 | 185.0 | GO:0070540 | stearic acid binding(GO:0070540) |
| 42.7 | 128.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 40.6 | 121.7 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 23.9 | 95.7 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 20.7 | 82.9 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 20.2 | 81.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 18.0 | 71.9 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 15.8 | 95.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 7.2 | 35.9 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 7.1 | 63.9 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 6.7 | 20.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 6.4 | 51.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 6.0 | 60.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 5.8 | 64.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 4.8 | 85.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 4.5 | 72.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 4.0 | 76.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 3.9 | 46.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 3.8 | 95.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 3.6 | 145.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 3.5 | 584.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 3.4 | 13.7 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 3.3 | 39.4 | GO:0019864 | IgG binding(GO:0019864) |
| 3.1 | 28.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 2.9 | 14.5 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 2.9 | 28.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 2.7 | 29.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 2.0 | 12.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 1.6 | 29.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 1.6 | 17.8 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 1.4 | 15.7 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 1.4 | 37.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 1.4 | 51.9 | GO:0071949 | FAD binding(GO:0071949) |
| 1.4 | 9.6 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 1.3 | 48.4 | GO:0016712 | arachidonic acid epoxygenase activity(GO:0008392) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 1.0 | 236.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.9 | 10.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.9 | 2.6 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.8 | 36.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.8 | 2.5 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.7 | 4.8 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.6 | 77.7 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.6 | 52.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.5 | 68.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.3 | 43.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.3 | 36.6 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.3 | 25.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.2 | 30.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.2 | 6.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 44.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.2 | 7.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 8.6 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.2 | 15.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 0.5 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 3.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 29.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 13.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 22.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 56.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 6.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 8.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 32.4 | GO:0016773 | phosphotransferase activity, alcohol group as acceptor(GO:0016773) |
| 0.0 | 0.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 2.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 17.1 | GO:0032403 | protein complex binding(GO:0032403) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 432.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 3.2 | 112.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.5 | 72.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.9 | 254.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.8 | 36.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.7 | 51.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.6 | 35.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.4 | 29.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 12.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 37.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.3 | 19.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 20.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 5.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 21.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 26.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.1 | 48.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 10.4 | 177.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 9.8 | 108.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 7.0 | 76.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 6.4 | 51.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 6.0 | 210.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 4.8 | 101.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 4.6 | 59.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 3.8 | 82.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 3.7 | 85.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 2.4 | 132.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 2.3 | 95.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 2.3 | 67.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.8 | 30.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.4 | 20.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.4 | 29.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 1.2 | 35.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 1.0 | 14.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.8 | 16.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.7 | 24.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 64.4 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.5 | 135.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 3.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 6.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |