Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
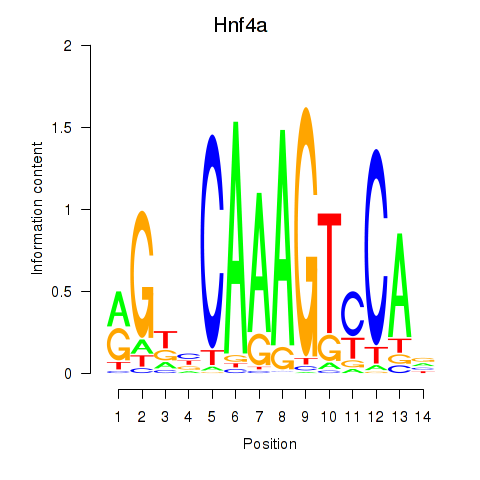
Results for Hnf4a
Z-value: 8.01
Transcription factors associated with Hnf4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hnf4a
|
ENSMUSG00000017950.17 | Hnf4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hnf4a | mm39_v1_chr2_+_163389068_163389108 | 0.93 | 1.1e-32 | Click! |
Activity profile of Hnf4a motif
Sorted Z-values of Hnf4a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hnf4a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_55995912 | 187.08 |
ENSMUST00000001497.9
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B |
| chr3_+_122688721 | 151.91 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr2_+_172994841 | 143.36 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chrX_+_10118544 | 131.50 |
ENSMUST00000049910.13
|
Otc
|
ornithine transcarbamylase |
| chrX_+_10118600 | 112.51 |
ENSMUST00000115528.3
|
Otc
|
ornithine transcarbamylase |
| chr19_-_39729431 | 110.01 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr10_-_128796834 | 106.31 |
ENSMUST00000026398.5
|
Mettl7b
|
methyltransferase like 7B |
| chr11_+_101258368 | 102.74 |
ENSMUST00000019469.3
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr15_-_82678490 | 101.55 |
ENSMUST00000006094.6
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26 |
| chr15_+_98997293 | 99.29 |
ENSMUST00000061295.7
|
Dnajc22
|
DnaJ heat shock protein family (Hsp40) member C22 |
| chr19_-_39801188 | 91.44 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr2_-_25390625 | 91.21 |
ENSMUST00000040042.11
|
C8g
|
complement component 8, gamma polypeptide |
| chr15_+_82336535 | 91.02 |
ENSMUST00000089129.7
ENSMUST00000229313.2 ENSMUST00000231136.2 |
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9 |
| chr3_+_89043879 | 86.11 |
ENSMUST00000107482.10
ENSMUST00000127058.2 |
Pklr
|
pyruvate kinase liver and red blood cell |
| chr3_+_89043440 | 84.26 |
ENSMUST00000047111.13
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chr18_+_21205386 | 83.43 |
ENSMUST00000082235.5
|
Mep1b
|
meprin 1 beta |
| chr10_-_25412010 | 82.74 |
ENSMUST00000179685.3
|
Smlr1
|
small leucine-rich protein 1 |
| chr4_-_62005498 | 81.02 |
ENSMUST00000107488.4
ENSMUST00000107472.8 ENSMUST00000084531.11 |
Mup3
|
major urinary protein 3 |
| chr3_+_59939175 | 80.74 |
ENSMUST00000029325.5
|
Aadac
|
arylacetamide deacetylase |
| chr9_+_46151994 | 80.53 |
ENSMUST00000034585.7
|
Apoa4
|
apolipoprotein A-IV |
| chr11_-_77784922 | 80.29 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr4_+_141473983 | 79.32 |
ENSMUST00000038161.5
|
Agmat
|
agmatine ureohydrolase (agmatinase) |
| chr16_+_37400500 | 79.01 |
ENSMUST00000160847.2
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr15_+_102058936 | 78.60 |
ENSMUST00000023806.14
|
Soat2
|
sterol O-acyltransferase 2 |
| chr16_+_37400590 | 78.26 |
ENSMUST00000159787.8
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr6_+_71176811 | 77.37 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr3_-_10400710 | 76.86 |
ENSMUST00000078748.4
|
Slc10a5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5 |
| chr19_-_39875192 | 76.73 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr7_-_135130374 | 76.00 |
ENSMUST00000053716.8
|
Clrn3
|
clarin 3 |
| chr9_-_46146928 | 74.46 |
ENSMUST00000118649.8
|
Apoc3
|
apolipoprotein C-III |
| chr19_-_40062174 | 74.18 |
ENSMUST00000048959.5
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
| chr1_+_139429430 | 73.33 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr7_+_44114815 | 73.26 |
ENSMUST00000035929.11
ENSMUST00000146128.8 |
Aspdh
|
aspartate dehydrogenase domain containing |
| chr19_+_39275518 | 72.43 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr5_-_104125270 | 71.73 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr15_+_10358611 | 70.71 |
ENSMUST00000110541.8
ENSMUST00000110542.8 |
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr15_-_82648376 | 70.63 |
ENSMUST00000055721.6
|
Cyp2d40
|
cytochrome P450, family 2, subfamily d, polypeptide 40 |
| chr5_-_104125192 | 69.40 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr19_+_12610668 | 68.86 |
ENSMUST00000044976.12
|
Glyat
|
glycine-N-acyltransferase |
| chr19_+_12610870 | 68.60 |
ENSMUST00000119960.2
|
Glyat
|
glycine-N-acyltransferase |
| chr7_-_19410749 | 68.20 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
| chr1_+_171052623 | 67.82 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr7_+_119773070 | 67.65 |
ENSMUST00000033201.7
|
Anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr19_+_39980868 | 65.17 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr19_+_44980565 | 63.99 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr19_-_10581622 | 63.89 |
ENSMUST00000037678.7
|
Tkfc
|
triokinase, FMN cyclase |
| chr19_-_39637489 | 63.08 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr15_-_82291372 | 61.71 |
ENSMUST00000230198.2
ENSMUST00000230248.2 ENSMUST00000072776.5 ENSMUST00000229911.2 |
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr15_+_10358643 | 61.21 |
ENSMUST00000022858.8
|
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr3_+_118355778 | 61.20 |
ENSMUST00000039177.12
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr6_-_142418801 | 60.44 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
| chr11_+_115768323 | 60.17 |
ENSMUST00000222123.2
|
Myo15b
|
myosin XVB |
| chr5_-_104125226 | 59.75 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr12_-_103871146 | 58.74 |
ENSMUST00000074051.6
|
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1C |
| chr7_-_105249308 | 58.39 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr3_+_96737385 | 58.31 |
ENSMUST00000058865.14
|
Pdzk1
|
PDZ domain containing 1 |
| chr19_-_8382424 | 57.00 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr7_+_44114857 | 56.27 |
ENSMUST00000135624.2
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr17_-_35081456 | 56.09 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr17_-_43813664 | 55.78 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr7_+_143027473 | 54.96 |
ENSMUST00000052348.12
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr17_-_35081129 | 53.84 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr19_+_38995463 | 52.15 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr11_-_75330302 | 51.97 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr1_+_171041583 | 51.84 |
ENSMUST00000111328.8
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr2_-_27138347 | 51.62 |
ENSMUST00000139312.8
|
Sardh
|
sarcosine dehydrogenase |
| chr4_-_60697274 | 51.05 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr5_-_115109118 | 50.99 |
ENSMUST00000031535.12
|
Hnf1a
|
HNF1 homeobox A |
| chr11_-_75330415 | 50.89 |
ENSMUST00000128330.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr11_+_69945157 | 50.80 |
ENSMUST00000108585.9
ENSMUST00000018699.13 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr7_-_25315299 | 50.13 |
ENSMUST00000098663.4
ENSMUST00000238895.2 |
Erich4
|
glutamate rich 4 |
| chr6_-_128503666 | 49.86 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr18_-_32170012 | 49.28 |
ENSMUST00000134663.2
|
Myo7b
|
myosin VIIB |
| chr1_+_165958036 | 48.75 |
ENSMUST00000166860.2
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr9_+_44584523 | 48.59 |
ENSMUST00000034609.11
ENSMUST00000071219.12 |
Treh
|
trehalase (brush-border membrane glycoprotein) |
| chr1_+_165957784 | 48.10 |
ENSMUST00000060833.14
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr1_+_157353696 | 47.79 |
ENSMUST00000111700.8
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr16_-_30086317 | 46.85 |
ENSMUST00000064856.9
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr4_+_118384426 | 46.56 |
ENSMUST00000030261.6
|
2610528J11Rik
|
RIKEN cDNA 2610528J11 gene |
| chr4_-_94538329 | 46.19 |
ENSMUST00000107101.2
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr8_-_93956143 | 45.62 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr3_+_118355811 | 45.52 |
ENSMUST00000149101.3
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr15_-_76501525 | 45.17 |
ENSMUST00000230977.2
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr15_+_3300249 | 44.68 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr8_+_13087805 | 44.32 |
ENSMUST00000128418.8
ENSMUST00000152034.2 |
F10
|
coagulation factor X |
| chr1_+_171041539 | 43.69 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr4_+_118384183 | 43.00 |
ENSMUST00000106367.8
|
2610528J11Rik
|
RIKEN cDNA 2610528J11 gene |
| chrX_-_16683578 | 42.45 |
ENSMUST00000040820.13
|
Maob
|
monoamine oxidase B |
| chr3_-_121608809 | 42.30 |
ENSMUST00000197383.5
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr19_-_8109346 | 41.55 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr2_-_119060330 | 41.17 |
ENSMUST00000110820.3
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr3_-_121608859 | 41.06 |
ENSMUST00000029770.8
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr2_-_155424576 | 40.75 |
ENSMUST00000126322.8
|
Gss
|
glutathione synthetase |
| chr5_-_137202790 | 40.69 |
ENSMUST00000041226.11
|
Muc3
|
mucin 3, intestinal |
| chr2_-_119060366 | 40.63 |
ENSMUST00000076084.6
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr1_+_165957909 | 40.17 |
ENSMUST00000166159.2
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr1_-_90897329 | 39.85 |
ENSMUST00000130042.2
ENSMUST00000027529.12 |
Rab17
|
RAB17, member RAS oncogene family |
| chr11_+_76795346 | 39.69 |
ENSMUST00000072633.4
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr8_+_13087292 | 39.59 |
ENSMUST00000063820.12
ENSMUST00000033821.11 |
F10
|
coagulation factor X |
| chr9_+_62765362 | 38.79 |
ENSMUST00000213643.2
ENSMUST00000034777.14 ENSMUST00000163820.3 ENSMUST00000215870.2 ENSMUST00000214633.2 ENSMUST00000215968.2 |
Calml4
|
calmodulin-like 4 |
| chr17_+_37209002 | 38.65 |
ENSMUST00000078438.5
|
Trim31
|
tripartite motif-containing 31 |
| chr7_-_45679703 | 38.59 |
ENSMUST00000002850.8
|
Abcc6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
| chr5_+_90708962 | 38.56 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr9_-_121745354 | 38.42 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr10_+_75407356 | 37.87 |
ENSMUST00000143226.8
ENSMUST00000124259.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr1_-_136888118 | 37.78 |
ENSMUST00000192357.6
ENSMUST00000027649.14 |
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr2_+_58645189 | 37.66 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr1_+_175459559 | 37.58 |
ENSMUST00000040250.15
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr16_+_33614378 | 36.75 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr2_-_110136074 | 36.70 |
ENSMUST00000046233.9
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr6_-_125357756 | 36.55 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr2_-_69172944 | 36.21 |
ENSMUST00000102709.8
ENSMUST00000102710.10 ENSMUST00000180142.2 |
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr16_+_33614715 | 35.84 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr12_+_59177780 | 35.72 |
ENSMUST00000177460.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr11_+_76795292 | 35.67 |
ENSMUST00000142166.8
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr9_-_35030479 | 35.13 |
ENSMUST00000213526.2
ENSMUST00000215089.2 |
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr1_+_185187000 | 35.12 |
ENSMUST00000061093.7
|
Slc30a10
|
solute carrier family 30, member 10 |
| chr1_-_87029312 | 35.01 |
ENSMUST00000113270.3
|
Alpi
|
alkaline phosphatase, intestinal |
| chr7_+_30193047 | 34.25 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr2_-_13496624 | 34.04 |
ENSMUST00000091436.7
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr4_+_95467653 | 33.40 |
ENSMUST00000043335.11
|
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr12_+_37291728 | 33.35 |
ENSMUST00000160768.8
|
Agmo
|
alkylglycerol monooxygenase |
| chr10_+_79658392 | 33.08 |
ENSMUST00000219305.2
ENSMUST00000046833.5 ENSMUST00000218687.2 |
Misp
|
mitotic spindle positioning |
| chr17_-_34822649 | 32.45 |
ENSMUST00000015622.8
|
Rnf5
|
ring finger protein 5 |
| chr17_-_46749370 | 32.20 |
ENSMUST00000087012.7
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr1_+_87983099 | 32.17 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr19_-_39451509 | 32.10 |
ENSMUST00000035488.3
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr15_+_82439273 | 31.93 |
ENSMUST00000229103.2
ENSMUST00000068861.8 ENSMUST00000229904.2 |
Cyp2d12
|
cytochrome P450, family 2, subfamily d, polypeptide 12 |
| chr7_+_28533279 | 31.59 |
ENSMUST00000208971.2
ENSMUST00000066723.15 |
Lgals4
|
lectin, galactose binding, soluble 4 |
| chr1_+_175459735 | 31.58 |
ENSMUST00000097458.4
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr8_-_122671588 | 31.53 |
ENSMUST00000057653.8
|
Car5a
|
carbonic anhydrase 5a, mitochondrial |
| chr12_+_37292029 | 31.06 |
ENSMUST00000160390.2
|
Agmo
|
alkylglycerol monooxygenase |
| chr8_+_13076024 | 30.75 |
ENSMUST00000033820.4
|
F7
|
coagulation factor VII |
| chr2_+_58644922 | 30.65 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr4_+_95467701 | 30.31 |
ENSMUST00000150830.2
ENSMUST00000134012.9 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr11_+_118878999 | 30.15 |
ENSMUST00000092373.13
ENSMUST00000106273.3 |
Enpp7
|
ectonucleotide pyrophosphatase/phosphodiesterase 7 |
| chr12_+_37291632 | 29.84 |
ENSMUST00000049874.14
|
Agmo
|
alkylglycerol monooxygenase |
| chr1_-_82746169 | 29.78 |
ENSMUST00000027331.3
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr11_+_73090270 | 29.50 |
ENSMUST00000006105.7
|
Shpk
|
sedoheptulokinase |
| chr15_-_82278223 | 29.20 |
ENSMUST00000170255.2
|
Cyp2d11
|
cytochrome P450, family 2, subfamily d, polypeptide 11 |
| chr19_-_44396092 | 28.72 |
ENSMUST00000041331.4
|
Scd1
|
stearoyl-Coenzyme A desaturase 1 |
| chr19_-_44396269 | 28.29 |
ENSMUST00000235741.2
|
Scd1
|
stearoyl-Coenzyme A desaturase 1 |
| chr17_+_34596098 | 28.16 |
ENSMUST00000080254.7
|
Btnl1
|
butyrophilin-like 1 |
| chr6_-_136758716 | 28.15 |
ENSMUST00000078095.11
ENSMUST00000032338.10 |
Gucy2c
|
guanylate cyclase 2c |
| chr11_+_83741657 | 27.81 |
ENSMUST00000021016.10
|
Hnf1b
|
HNF1 homeobox B |
| chr8_-_13250535 | 27.28 |
ENSMUST00000165605.4
ENSMUST00000209691.2 ENSMUST00000211128.2 ENSMUST00000210317.2 |
Grtp1
|
GH regulated TBC protein 1 |
| chr4_+_134124691 | 26.94 |
ENSMUST00000105870.8
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr9_-_44162762 | 26.70 |
ENSMUST00000034618.6
|
Pdzd3
|
PDZ domain containing 3 |
| chr9_-_106035308 | 26.30 |
ENSMUST00000159809.2
ENSMUST00000162562.2 ENSMUST00000036382.13 |
Glyctk
|
glycerate kinase |
| chr11_+_83741689 | 26.22 |
ENSMUST00000108114.9
|
Hnf1b
|
HNF1 homeobox B |
| chr10_+_18720760 | 26.14 |
ENSMUST00000019998.9
|
Perp
|
PERP, TP53 apoptosis effector |
| chr5_+_31454787 | 26.07 |
ENSMUST00000201166.4
ENSMUST00000072228.9 |
Gckr
|
glucokinase regulatory protein |
| chr2_+_156928962 | 25.81 |
ENSMUST00000166140.8
|
Tldc2
|
TBC/LysM associated domain containing 2 |
| chr4_-_94538370 | 25.79 |
ENSMUST00000053419.9
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr12_+_111237523 | 25.09 |
ENSMUST00000021707.8
|
Amn
|
amnionless |
| chr1_+_87983189 | 24.99 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr11_+_78389913 | 24.90 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr4_-_62069046 | 24.87 |
ENSMUST00000077719.4
|
Mup21
|
major urinary protein 21 |
| chr19_-_11058452 | 23.91 |
ENSMUST00000025636.8
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr10_+_75402090 | 23.88 |
ENSMUST00000129232.8
ENSMUST00000143792.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr9_-_110818679 | 23.49 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chr17_-_35100980 | 23.16 |
ENSMUST00000152417.8
ENSMUST00000146299.8 |
C2
Gm20547
|
complement component 2 (within H-2S) predicted gene 20547 |
| chr2_-_134396268 | 22.89 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr3_-_151871867 | 22.80 |
ENSMUST00000046614.10
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr11_+_7147779 | 22.79 |
ENSMUST00000020704.8
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr7_+_122813999 | 22.48 |
ENSMUST00000131933.8
|
Slc5a11
|
solute carrier family 5 (sodium/glucose cotransporter), member 11 |
| chr11_-_61157986 | 22.23 |
ENSMUST00000066277.10
ENSMUST00000074127.14 ENSMUST00000108715.3 |
Aldh3a2
|
aldehyde dehydrogenase family 3, subfamily A2 |
| chr17_-_46956054 | 22.19 |
ENSMUST00000003642.7
|
Klc4
|
kinesin light chain 4 |
| chr9_-_106035332 | 22.16 |
ENSMUST00000112543.9
|
Glyctk
|
glycerate kinase |
| chr7_+_122814031 | 22.15 |
ENSMUST00000033035.13
|
Slc5a11
|
solute carrier family 5 (sodium/glucose cotransporter), member 11 |
| chr15_-_89064936 | 21.95 |
ENSMUST00000109331.9
|
Plxnb2
|
plexin B2 |
| chr6_-_85114725 | 21.94 |
ENSMUST00000174769.2
ENSMUST00000174286.3 ENSMUST00000045986.8 |
Spr
|
sepiapterin reductase |
| chr17_-_37178079 | 21.90 |
ENSMUST00000025329.13
ENSMUST00000174195.8 |
Trim15
|
tripartite motif-containing 15 |
| chr5_+_137568982 | 21.46 |
ENSMUST00000196471.5
ENSMUST00000198783.5 |
Tfr2
|
transferrin receptor 2 |
| chr17_-_46749320 | 21.29 |
ENSMUST00000233575.2
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr15_-_76004395 | 21.11 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1 |
| chr10_-_128755127 | 20.89 |
ENSMUST00000149961.2
ENSMUST00000026406.14 |
Rdh5
|
retinol dehydrogenase 5 |
| chr6_-_86742789 | 20.73 |
ENSMUST00000123732.4
|
Anxa4
|
annexin A4 |
| chr2_-_130480014 | 20.16 |
ENSMUST00000089561.10
ENSMUST00000110260.8 |
Lzts3
|
leucine zipper, putative tumor suppressor family member 3 |
| chr8_+_3637785 | 20.10 |
ENSMUST00000171962.3
ENSMUST00000207712.2 ENSMUST00000207970.2 ENSMUST00000207533.2 ENSMUST00000208240.2 ENSMUST00000207432.2 ENSMUST00000207077.2 |
Camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr12_+_21366386 | 19.90 |
ENSMUST00000076813.8
ENSMUST00000221693.2 ENSMUST00000223345.2 ENSMUST00000222344.2 |
Iah1
|
isoamyl acetate-hydrolyzing esterase 1 homolog |
| chr13_+_55547498 | 19.75 |
ENSMUST00000057167.9
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1 |
| chr8_-_41469332 | 19.72 |
ENSMUST00000131965.2
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr15_-_82505132 | 19.54 |
ENSMUST00000109515.3
|
Cyp2d34
|
cytochrome P450, family 2, subfamily d, polypeptide 34 |
| chr7_-_99344779 | 19.34 |
ENSMUST00000137914.2
ENSMUST00000207090.2 ENSMUST00000208225.2 |
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr19_+_39049442 | 19.03 |
ENSMUST00000087236.5
|
Cyp2c65
|
cytochrome P450, family 2, subfamily c, polypeptide 65 |
| chr9_-_55419442 | 18.94 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr5_+_8943943 | 18.79 |
ENSMUST00000196067.2
|
Abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr17_-_34962823 | 18.72 |
ENSMUST00000069507.9
|
C4b
|
complement component 4B (Chido blood group) |
| chr15_-_78739717 | 18.15 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr6_+_72333209 | 17.69 |
ENSMUST00000206531.2
|
Tmem150a
|
transmembrane protein 150A |
| chr11_+_70410009 | 17.54 |
ENSMUST00000057685.3
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr6_-_86742847 | 17.54 |
ENSMUST00000113675.8
|
Anxa4
|
annexin A4 |
| chr10_+_127685785 | 17.36 |
ENSMUST00000077530.3
|
Rdh19
|
retinol dehydrogenase 19 |
| chr8_+_87350672 | 17.35 |
ENSMUST00000034141.18
ENSMUST00000122188.10 |
Lonp2
|
lon peptidase 2, peroxisomal |
| chr16_-_17745999 | 17.15 |
ENSMUST00000003622.16
|
Slc25a1
|
solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 |
| chr9_-_110576192 | 17.04 |
ENSMUST00000199791.2
|
Pth1r
|
parathyroid hormone 1 receptor |
| chrX_+_95498965 | 16.61 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr12_+_59177552 | 16.13 |
ENSMUST00000175877.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr2_+_110427643 | 15.70 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 81.3 | 244.0 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 44.0 | 131.9 | GO:0042853 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) |
| 35.8 | 143.4 | GO:0061402 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 35.6 | 106.7 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 26.8 | 80.5 | GO:0034443 | regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) multicellular organism lipid catabolic process(GO:0044240) |
| 26.8 | 80.3 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 23.8 | 142.7 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 23.4 | 116.9 | GO:1904970 | brush border assembly(GO:1904970) |
| 21.6 | 129.5 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 20.8 | 83.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 20.2 | 666.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 19.0 | 57.0 | GO:1903699 | tarsal gland development(GO:1903699) |
| 18.9 | 170.4 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 17.5 | 157.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 17.2 | 51.6 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 17.0 | 67.8 | GO:0046340 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) diacylglycerol catabolic process(GO:0046340) |
| 13.5 | 54.0 | GO:0061235 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 13.3 | 39.8 | GO:0002414 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 13.2 | 79.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 12.9 | 102.9 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 11.8 | 201.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 11.7 | 35.1 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 11.5 | 69.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 11.4 | 34.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 10.7 | 32.1 | GO:1903413 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 10.6 | 63.9 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 10.3 | 102.7 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 10.1 | 80.7 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 9.9 | 59.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 9.8 | 29.5 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 9.8 | 78.6 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 9.7 | 405.6 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 9.5 | 47.3 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 9.2 | 64.6 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 9.2 | 36.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 9.0 | 45.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 9.0 | 9.0 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 8.5 | 42.5 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 7.8 | 39.2 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 7.7 | 61.8 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 7.7 | 115.8 | GO:0015747 | urate transport(GO:0015747) |
| 7.5 | 22.5 | GO:0060464 | lung lobe formation(GO:0060464) diaphragm morphogenesis(GO:0060540) |
| 7.4 | 22.2 | GO:1903173 | sesquiterpenoid metabolic process(GO:0006714) phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 7.3 | 29.2 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 7.3 | 94.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 7.2 | 36.2 | GO:0046618 | drug export(GO:0046618) |
| 6.6 | 19.8 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 5.6 | 134.5 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 5.6 | 44.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 5.5 | 225.4 | GO:0050892 | intestinal absorption(GO:0050892) |
| 5.5 | 76.4 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 5.3 | 58.3 | GO:0015879 | carnitine transport(GO:0015879) |
| 5.2 | 26.1 | GO:0009750 | response to fructose(GO:0009750) |
| 5.1 | 30.7 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 5.1 | 51.0 | GO:0043691 | reverse cholesterol transport(GO:0043691) |
| 4.8 | 47.8 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 4.8 | 57.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 4.7 | 28.2 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 4.6 | 22.9 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 4.5 | 9.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 4.5 | 72.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 4.4 | 21.9 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 4.3 | 38.3 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 4.0 | 178.0 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 3.9 | 23.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 3.8 | 30.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 3.7 | 29.7 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 3.7 | 21.9 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 3.5 | 14.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 3.4 | 17.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 3.3 | 13.3 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 3.3 | 22.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 3.1 | 37.8 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 2.9 | 40.6 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 2.8 | 14.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 2.8 | 8.4 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 2.7 | 26.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 2.5 | 12.5 | GO:0015808 | L-alanine transport(GO:0015808) |
| 2.4 | 40.8 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 2.3 | 6.8 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 2.2 | 6.6 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 2.1 | 23.4 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 2.1 | 24.9 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 2.0 | 21.5 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 1.9 | 13.1 | GO:0015862 | uridine transport(GO:0015862) |
| 1.8 | 32.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 1.8 | 18.0 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 1.8 | 16.1 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 1.8 | 10.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 1.7 | 26.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 1.7 | 8.6 | GO:0090366 | DNA cytosine deamination(GO:0070383) positive regulation of mRNA modification(GO:0090366) |
| 1.7 | 69.0 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 1.7 | 83.4 | GO:1901998 | toxin transport(GO:1901998) |
| 1.6 | 11.5 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 1.6 | 30.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 1.5 | 191.5 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 1.5 | 37.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 1.5 | 55.0 | GO:0015893 | drug transport(GO:0015893) |
| 1.4 | 89.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 1.4 | 5.7 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 1.4 | 4.2 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 1.3 | 9.0 | GO:0001757 | somite specification(GO:0001757) |
| 1.3 | 8.9 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 1.3 | 26.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 1.2 | 35.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 1.2 | 11.6 | GO:0098706 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 1.1 | 2.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 1.1 | 5.3 | GO:0097459 | iron ion import into cell(GO:0097459) iron ion import across plasma membrane(GO:0098711) |
| 1.0 | 60.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 1.0 | 7.8 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 1.0 | 8.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.9 | 19.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.9 | 23.5 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.9 | 7.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.8 | 72.0 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.8 | 25.8 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.7 | 3.5 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.7 | 49.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.7 | 2.8 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.7 | 3.9 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.7 | 28.2 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.6 | 5.2 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.6 | 13.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.6 | 19.7 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.6 | 63.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.6 | 10.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.6 | 3.4 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.6 | 68.3 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.5 | 14.1 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.5 | 31.5 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.5 | 5.3 | GO:0070814 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.5 | 17.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.5 | 5.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.5 | 86.8 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.5 | 6.7 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.5 | 2.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.4 | 78.3 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.4 | 9.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.4 | 7.1 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.4 | 4.9 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.4 | 20.0 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.3 | 10.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.3 | 27.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.3 | 3.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.3 | 4.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 3.4 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.3 | 79.3 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.3 | 57.7 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.3 | 50.8 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.2 | 1.3 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.2 | 1.9 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 4.9 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.2 | 42.8 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 6.2 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 4.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 59.2 | GO:0032259 | methylation(GO:0032259) |
| 0.1 | 1.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 19.9 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 0.8 | GO:0019243 | methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 27.7 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.1 | 10.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 11.2 | GO:0045861 | negative regulation of proteolysis(GO:0045861) |
| 0.1 | 3.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 15.1 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 6.7 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.0 | 2.0 | GO:0007596 | blood coagulation(GO:0007596) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.8 | 229.3 | GO:0045179 | apical cortex(GO:0045179) |
| 14.0 | 210.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 11.4 | 102.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 10.1 | 91.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 7.0 | 35.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 6.8 | 34.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 6.7 | 26.6 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 6.2 | 68.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 4.7 | 80.5 | GO:0042627 | chylomicron(GO:0042627) |
| 3.8 | 68.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 3.6 | 407.7 | GO:0005811 | lipid particle(GO:0005811) |
| 3.6 | 24.9 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 3.5 | 101.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 3.2 | 22.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 2.0 | 69.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 2.0 | 32.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 1.8 | 54.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 1.7 | 13.5 | GO:0008091 | spectrin(GO:0008091) |
| 1.7 | 21.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.6 | 190.5 | GO:0005902 | microvillus(GO:0005902) |
| 1.5 | 20.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.5 | 10.5 | GO:0070695 | FHF complex(GO:0070695) |
| 1.4 | 60.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 1.4 | 51.0 | GO:0045120 | pronucleus(GO:0045120) |
| 1.3 | 9.0 | GO:1990357 | terminal web(GO:1990357) |
| 1.2 | 250.6 | GO:0005903 | brush border(GO:0005903) |
| 1.2 | 28.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 1.0 | 29.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.9 | 342.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.9 | 235.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.8 | 143.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.8 | 5.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.6 | 311.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.5 | 1053.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.5 | 83.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.5 | 40.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.5 | 11.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.4 | 35.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.4 | 1093.6 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.3 | 31.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.3 | 54.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.2 | 38.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.2 | 20.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.2 | 266.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.2 | 214.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.2 | 16.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 118.4 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 66.7 | GO:0005768 | endosome(GO:0005768) |
| 0.1 | 7.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 3.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 6.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 111.3 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.1 | 2.4 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 127.7 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 6.9 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 61.0 | 244.0 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 47.8 | 143.4 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 35.6 | 106.7 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 35.6 | 142.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 34.7 | 312.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 34.4 | 137.5 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 34.1 | 170.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 31.6 | 126.3 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 31.4 | 94.3 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 25.7 | 102.7 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 22.3 | 133.6 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 22.0 | 131.9 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 21.3 | 63.9 | GO:0004371 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 21.2 | 63.7 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 20.1 | 60.4 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 17.2 | 51.6 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 17.1 | 68.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 16.3 | 440.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 13.4 | 80.5 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 13.4 | 80.3 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 12.5 | 249.8 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 11.4 | 68.3 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 11.3 | 45.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 9.7 | 29.2 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 9.7 | 58.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 9.6 | 76.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 9.5 | 57.0 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 7.8 | 23.4 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 7.3 | 95.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 7.3 | 58.3 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 7.2 | 79.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 6.8 | 115.8 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 6.3 | 69.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 5.9 | 172.0 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 5.5 | 160.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 5.5 | 49.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 5.3 | 32.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 5.1 | 61.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 5.1 | 91.2 | GO:0019841 | retinol binding(GO:0019841) |
| 5.0 | 75.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 5.0 | 35.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 5.0 | 35.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 4.9 | 34.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 4.7 | 14.1 | GO:0047223 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 4.7 | 164.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 4.4 | 13.3 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 4.4 | 30.6 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 4.3 | 17.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 4.3 | 21.5 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 4.2 | 12.5 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 4.0 | 19.8 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 3.8 | 80.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 3.8 | 49.9 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 3.8 | 26.7 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 3.5 | 14.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 3.4 | 17.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 3.4 | 40.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 3.3 | 46.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 3.0 | 30.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 2.7 | 29.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 2.7 | 10.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 2.6 | 23.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 2.6 | 26.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 2.5 | 44.7 | GO:0008430 | selenium binding(GO:0008430) |
| 2.3 | 18.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.3 | 55.0 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 2.3 | 22.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 2.3 | 9.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 2.2 | 53.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 2.2 | 8.7 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 2.0 | 82.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 1.9 | 26.9 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 1.9 | 7.7 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 1.9 | 40.8 | GO:0016594 | glycine binding(GO:0016594) |
| 1.8 | 32.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 1.8 | 5.3 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 1.7 | 15.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 1.7 | 38.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 1.7 | 5.2 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 1.7 | 8.6 | GO:0004131 | cytosine deaminase activity(GO:0004131) |
| 1.7 | 22.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 1.7 | 24.8 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 1.6 | 55.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 1.6 | 11.5 | GO:0050294 | alcohol sulfotransferase activity(GO:0004027) steroid sulfotransferase activity(GO:0050294) |
| 1.6 | 87.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 1.5 | 28.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 1.5 | 11.6 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.4 | 57.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.4 | 16.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.3 | 25.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 1.3 | 34.2 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 1.3 | 22.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 1.1 | 138.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 1.1 | 247.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 1.0 | 20.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 1.0 | 7.8 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.9 | 71.8 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.9 | 141.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.8 | 8.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.8 | 38.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.7 | 51.0 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.7 | 3.5 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.7 | 6.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.6 | 6.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.6 | 4.9 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.5 | 37.8 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.5 | 62.4 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.5 | 72.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.5 | 2.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.5 | 36.7 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.5 | 36.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.5 | 5.9 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.4 | 5.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.4 | 6.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.4 | 12.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.4 | 47.3 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.4 | 40.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.3 | 54.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.3 | 1.9 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 6.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 38.6 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.3 | 18.9 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.3 | 74.4 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.3 | 4.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 158.9 | GO:0016491 | oxidoreductase activity(GO:0016491) |
| 0.2 | 19.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.2 | 16.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 9.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 4.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 16.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 27.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 5.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 20.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.8 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 3.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 3.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 2.4 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 3.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 53.7 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 4.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 3.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 544.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 3.6 | 98.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 1.6 | 452.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 1.6 | 326.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 1.1 | 24.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.8 | 42.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.7 | 91.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.6 | 20.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.4 | 26.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 29.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 22.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.3 | 22.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 14.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.2 | 18.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 7.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 6.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 4.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.3 | 306.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 12.1 | 48.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 9.9 | 197.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 9.8 | 157.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 9.5 | 133.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 9.0 | 143.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 8.3 | 108.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 6.3 | 221.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 5.4 | 91.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 4.9 | 69.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 4.6 | 91.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 4.4 | 79.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 3.6 | 36.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 3.4 | 30.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 3.2 | 67.8 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 3.0 | 39.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 3.0 | 38.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 2.8 | 72.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 2.6 | 251.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 2.2 | 35.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 2.1 | 119.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 1.9 | 432.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 1.8 | 47.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 1.6 | 67.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 1.3 | 22.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 1.3 | 23.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 1.2 | 29.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 1.2 | 21.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 1.1 | 16.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 1.1 | 22.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.1 | 18.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.0 | 95.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.9 | 84.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.8 | 94.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.8 | 42.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.8 | 30.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.8 | 102.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.7 | 30.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.7 | 22.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 44.6 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.5 | 9.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.5 | 14.1 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.4 | 6.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 3.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 19.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 13.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 5.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 13.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 12.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 4.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 19.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 27.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 3.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.2 | 1.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 13.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 5.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 3.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |