Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
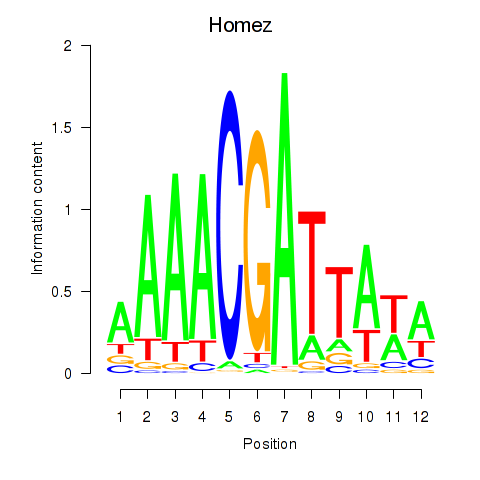
Results for Homez
Z-value: 0.83
Transcription factors associated with Homez
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Homez
|
ENSMUSG00000057156.11 | Homez |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Homez | mm39_v1_chr14_-_55101505_55101615 | 0.47 | 2.7e-05 | Click! |
Activity profile of Homez motif
Sorted Z-values of Homez motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Homez
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_124746510 | 3.35 |
ENSMUST00000149652.2
ENSMUST00000112476.8 ENSMUST00000004378.15 |
Eno2
|
enolase 2, gamma neuronal |
| chr1_-_52271455 | 3.14 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr6_+_71350411 | 3.06 |
ENSMUST00000066747.14
|
Cd8a
|
CD8 antigen, alpha chain |
| chr6_+_71350519 | 2.87 |
ENSMUST00000172321.3
|
Cd8a
|
CD8 antigen, alpha chain |
| chr16_-_48592372 | 2.82 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr5_-_148931957 | 2.74 |
ENSMUST00000147473.6
|
Gm42791
|
predicted gene 42791 |
| chr17_+_9142941 | 2.73 |
ENSMUST00000149440.8
|
Pde10a
|
phosphodiesterase 10A |
| chr16_-_48592319 | 2.58 |
ENSMUST00000239408.2
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr2_+_112097087 | 2.55 |
ENSMUST00000110987.9
ENSMUST00000028549.14 |
Slc12a6
|
solute carrier family 12, member 6 |
| chr12_+_105420089 | 2.52 |
ENSMUST00000178224.2
|
D430019H16Rik
|
RIKEN cDNA D430019H16 gene |
| chr6_+_78347636 | 2.41 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr15_+_100052260 | 2.39 |
ENSMUST00000023768.14
|
Dip2b
|
disco interacting protein 2 homolog B |
| chr4_-_117035922 | 2.39 |
ENSMUST00000153953.2
ENSMUST00000106436.8 |
Kif2c
|
kinesin family member 2C |
| chr1_-_80642969 | 2.36 |
ENSMUST00000190983.7
ENSMUST00000191449.2 |
Dock10
|
dedicator of cytokinesis 10 |
| chrY_-_1286623 | 2.31 |
ENSMUST00000091190.12
|
Ddx3y
|
DEAD box helicase 3, Y-linked |
| chr15_-_98118858 | 2.31 |
ENSMUST00000142443.8
ENSMUST00000170618.8 |
Gm44579
Olfr287
|
predicted gene 44579 olfactory receptor 287 |
| chr1_+_72323526 | 2.23 |
ENSMUST00000027380.12
ENSMUST00000141783.2 |
Tmem169
|
transmembrane protein 169 |
| chr16_-_19703014 | 2.13 |
ENSMUST00000100083.5
ENSMUST00000231564.2 |
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr13_+_117356808 | 2.09 |
ENSMUST00000022242.9
|
Emb
|
embigin |
| chr13_-_117161921 | 2.03 |
ENSMUST00000223949.2
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr4_-_150736554 | 1.94 |
ENSMUST00000117997.2
ENSMUST00000037827.10 |
Slc45a1
|
solute carrier family 45, member 1 |
| chr19_-_41732104 | 1.92 |
ENSMUST00000025993.10
|
Slit1
|
slit guidance ligand 1 |
| chr6_-_78445846 | 1.90 |
ENSMUST00000032089.3
|
Reg3g
|
regenerating islet-derived 3 gamma |
| chr4_-_62389098 | 1.90 |
ENSMUST00000135811.2
ENSMUST00000120095.8 ENSMUST00000030087.14 ENSMUST00000107452.8 ENSMUST00000155522.8 |
Wdr31
|
WD repeat domain 31 |
| chr11_+_58845502 | 1.85 |
ENSMUST00000108817.5
ENSMUST00000047697.12 |
H2aw
Trim17
|
H2A.W histone tripartite motif-containing 17 |
| chr13_-_117162041 | 1.81 |
ENSMUST00000022239.8
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr2_-_151586063 | 1.81 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr6_-_69282389 | 1.80 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr18_+_50164043 | 1.79 |
ENSMUST00000145726.2
ENSMUST00000128377.2 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr4_+_102617332 | 1.77 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chrX_+_72108393 | 1.73 |
ENSMUST00000060418.8
|
Pnma3
|
paraneoplastic antigen MA3 |
| chr9_-_75506406 | 1.71 |
ENSMUST00000215036.2
|
Tmod2
|
tropomodulin 2 |
| chr13_-_51723473 | 1.70 |
ENSMUST00000239056.2
ENSMUST00000223543.3 |
Shc3
|
src homology 2 domain-containing transforming protein C3 |
| chr13_-_8921732 | 1.69 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr18_-_67682312 | 1.64 |
ENSMUST00000224799.2
|
Spire1
|
spire type actin nucleation factor 1 |
| chr1_+_82702598 | 1.62 |
ENSMUST00000078332.13
ENSMUST00000073025.12 ENSMUST00000161648.8 ENSMUST00000160786.8 ENSMUST00000162003.8 |
Mff
|
mitochondrial fission factor |
| chr14_+_27598021 | 1.62 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr6_-_69415741 | 1.60 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chrX_+_165021897 | 1.60 |
ENSMUST00000112235.8
|
Gpm6b
|
glycoprotein m6b |
| chr14_-_20026761 | 1.57 |
ENSMUST00000161247.2
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr8_-_4829519 | 1.57 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr10_+_129219952 | 1.57 |
ENSMUST00000214064.2
|
Olfr784
|
olfactory receptor 784 |
| chr6_+_56691875 | 1.55 |
ENSMUST00000031805.11
ENSMUST00000177249.3 |
Avl9
|
AVL9 cell migration associated |
| chr5_+_37403098 | 1.55 |
ENSMUST00000031004.11
|
Crmp1
|
collapsin response mediator protein 1 |
| chr13_+_19369097 | 1.54 |
ENSMUST00000103554.5
|
Trgv4
|
T cell receptor gamma, variable 4 |
| chr1_+_152830720 | 1.53 |
ENSMUST00000043313.15
ENSMUST00000186621.2 |
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr6_+_41520737 | 1.52 |
ENSMUST00000103298.2
|
Trbj2-7
|
T cell receptor beta joining 2-7 |
| chr8_-_94825556 | 1.51 |
ENSMUST00000034206.6
|
Bbs2
|
Bardet-Biedl syndrome 2 (human) |
| chr13_+_43019718 | 1.48 |
ENSMUST00000131942.2
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr6_-_69800923 | 1.47 |
ENSMUST00000103368.3
|
Igkv5-43
|
immunoglobulin kappa chain variable 5-43 |
| chr10_+_25317309 | 1.47 |
ENSMUST00000217929.2
ENSMUST00000220121.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr13_-_97334859 | 1.46 |
ENSMUST00000022169.10
|
Hexb
|
hexosaminidase B |
| chr14_-_27230402 | 1.45 |
ENSMUST00000050480.8
ENSMUST00000223689.2 |
Ccdc66
|
coiled-coil domain containing 66 |
| chr1_-_153425791 | 1.44 |
ENSMUST00000041874.9
|
Npl
|
N-acetylneuraminate pyruvate lyase |
| chr19_-_36896999 | 1.44 |
ENSMUST00000238948.2
ENSMUST00000057337.9 |
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr4_-_139974062 | 1.43 |
ENSMUST00000039331.9
|
Igsf21
|
immunoglobulin superfamily, member 21 |
| chr5_-_148988110 | 1.42 |
ENSMUST00000110505.8
|
Hmgb1
|
high mobility group box 1 |
| chr14_+_54198389 | 1.41 |
ENSMUST00000103678.4
|
Trdv2-2
|
T cell receptor delta variable 2-2 |
| chr2_+_14828903 | 1.40 |
ENSMUST00000193800.6
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr7_-_12552764 | 1.38 |
ENSMUST00000108546.2
ENSMUST00000072222.8 |
Zfp329
|
zinc finger protein 329 |
| chr18_+_86729184 | 1.36 |
ENSMUST00000068423.10
|
Cbln2
|
cerebellin 2 precursor protein |
| chr19_-_4240984 | 1.35 |
ENSMUST00000045864.4
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr2_-_57942844 | 1.33 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein |
| chr12_-_79030250 | 1.33 |
ENSMUST00000070174.14
|
Tmem229b
|
transmembrane protein 229B |
| chr7_-_119078472 | 1.32 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr8_-_73059104 | 1.31 |
ENSMUST00000075602.8
|
Gm10282
|
predicted pseudogene 10282 |
| chr6_+_79995860 | 1.31 |
ENSMUST00000147663.8
ENSMUST00000128718.8 ENSMUST00000126005.8 ENSMUST00000133918.8 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr1_-_163822336 | 1.30 |
ENSMUST00000097493.10
ENSMUST00000045876.8 |
BC055324
|
cDNA sequence BC055324 |
| chr16_+_17307503 | 1.30 |
ENSMUST00000023448.15
|
Aifm3
|
apoptosis-inducing factor, mitochondrion-associated 3 |
| chr7_+_19927635 | 1.30 |
ENSMUST00000168984.2
|
Vmn1r95
|
vomeronasal 1 receptor, 95 |
| chr14_+_79689230 | 1.30 |
ENSMUST00000100359.3
ENSMUST00000226192.2 |
Kbtbd6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr16_+_11224481 | 1.29 |
ENSMUST00000122168.8
|
Snx29
|
sorting nexin 29 |
| chr6_-_144978557 | 1.28 |
ENSMUST00000136819.3
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr14_+_54113415 | 1.28 |
ENSMUST00000180938.3
|
Trav21-dv12
|
T cell receptor alpha variable 21-DV12 |
| chr4_-_126954878 | 1.28 |
ENSMUST00000136186.2
ENSMUST00000106099.8 ENSMUST00000106102.9 |
Zmym1
|
zinc finger, MYM domain containing 1 |
| chr10_+_128583734 | 1.27 |
ENSMUST00000163377.10
|
Pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr7_+_18915136 | 1.26 |
ENSMUST00000144054.8
ENSMUST00000141718.8 |
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr15_-_98676007 | 1.26 |
ENSMUST00000226655.2
ENSMUST00000228546.2 ENSMUST00000023732.12 ENSMUST00000226610.2 |
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr2_-_164120215 | 1.25 |
ENSMUST00000017142.3
|
Svs4
|
seminal vesicle secretory protein 4 |
| chr10_+_57660948 | 1.25 |
ENSMUST00000020024.12
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr17_+_21876498 | 1.24 |
ENSMUST00000039726.8
|
Zfp983
|
zinc finger protein 983 |
| chr9_-_15834052 | 1.24 |
ENSMUST00000217187.2
|
Fat3
|
FAT atypical cadherin 3 |
| chr12_-_114710326 | 1.23 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr18_-_78100610 | 1.22 |
ENSMUST00000170760.3
|
Siglec15
|
sialic acid binding Ig-like lectin 15 |
| chr15_+_102204691 | 1.21 |
ENSMUST00000064924.6
ENSMUST00000230212.2 ENSMUST00000229050.2 ENSMUST00000231104.2 |
Espl1
|
extra spindle pole bodies 1, separase |
| chr15_-_36792649 | 1.20 |
ENSMUST00000126184.2
|
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr3_-_50398027 | 1.19 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr8_-_46533321 | 1.18 |
ENSMUST00000177186.2
|
Snx25
|
sorting nexin 25 |
| chr6_+_120440159 | 1.18 |
ENSMUST00000002976.5
|
Il17ra
|
interleukin 17 receptor A |
| chr7_+_126549692 | 1.16 |
ENSMUST00000106335.8
ENSMUST00000146017.3 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr11_-_101917745 | 1.16 |
ENSMUST00000107167.2
ENSMUST00000062801.11 |
Mpp3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
| chr14_+_76192449 | 1.13 |
ENSMUST00000050120.4
|
Kctd4
|
potassium channel tetramerisation domain containing 4 |
| chr5_-_21990170 | 1.13 |
ENSMUST00000115193.8
ENSMUST00000115192.2 ENSMUST00000115195.8 ENSMUST00000030771.12 |
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chr2_-_92231598 | 1.12 |
ENSMUST00000050312.3
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr6_+_70648743 | 1.12 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4 |
| chr7_-_119078330 | 1.12 |
ENSMUST00000207460.2
|
Umod
|
uromodulin |
| chr6_+_103674695 | 1.11 |
ENSMUST00000205098.2
|
Chl1
|
cell adhesion molecule L1-like |
| chr7_-_81139550 | 1.10 |
ENSMUST00000238692.2
|
Ap3b2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr7_-_29750785 | 1.09 |
ENSMUST00000207072.2
ENSMUST00000207873.2 |
Zfp14
|
zinc finger protein 14 |
| chr17_+_66418525 | 1.09 |
ENSMUST00000072383.14
|
Washc1
|
WASH complex subunit 1 |
| chr12_-_103392039 | 1.08 |
ENSMUST00000110001.4
ENSMUST00000223233.2 ENSMUST00000044923.15 ENSMUST00000221211.2 |
Ddx24
|
DEAD box helicase 24 |
| chr7_-_29772226 | 1.07 |
ENSMUST00000183115.8
ENSMUST00000182919.8 ENSMUST00000183190.2 ENSMUST00000080834.15 |
Zfp82
|
zinc finger protein 82 |
| chr7_+_3352159 | 1.07 |
ENSMUST00000172109.4
|
Prkcg
|
protein kinase C, gamma |
| chr4_+_85123358 | 1.07 |
ENSMUST00000107188.10
|
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr8_+_21545063 | 1.07 |
ENSMUST00000098899.4
|
Defa23
|
defensin, alpha, 23 |
| chr6_-_85046430 | 1.06 |
ENSMUST00000160197.6
|
Exoc6b
|
exocyst complex component 6B |
| chr10_-_18110682 | 1.06 |
ENSMUST00000052648.9
ENSMUST00000080860.13 ENSMUST00000173243.8 |
Ccdc28a
|
coiled-coil domain containing 28A |
| chr8_+_117648474 | 1.06 |
ENSMUST00000034205.5
ENSMUST00000212775.2 |
Cenpn
|
centromere protein N |
| chr6_-_136918885 | 1.06 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr2_-_106804679 | 1.06 |
ENSMUST00000028536.13
|
Arl14ep
|
ADP-ribosylation factor-like 14 effector protein |
| chr1_-_180971763 | 1.05 |
ENSMUST00000027797.9
|
Nvl
|
nuclear VCP-like |
| chr19_-_46958001 | 1.05 |
ENSMUST00000235234.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr2_+_118865262 | 1.05 |
ENSMUST00000028796.2
|
Rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr18_-_12429048 | 1.04 |
ENSMUST00000234212.2
|
Ankrd29
|
ankyrin repeat domain 29 |
| chr2_-_86180622 | 1.04 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr11_+_120489358 | 1.03 |
ENSMUST00000093140.5
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr2_-_66240408 | 1.03 |
ENSMUST00000112366.8
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr16_+_95946591 | 1.01 |
ENSMUST00000023913.11
ENSMUST00000232832.2 ENSMUST00000233566.2 ENSMUST00000233273.2 |
Get1
|
guided entry of tail-anchored proteins factor 1 |
| chr18_+_38809771 | 1.01 |
ENSMUST00000134388.2
ENSMUST00000148850.8 |
9630014M24Rik
Arhgap26
|
RIKEN cDNA 9630014M24 gene Rho GTPase activating protein 26 |
| chr1_+_78635542 | 1.01 |
ENSMUST00000035779.15
|
Acsl3
|
acyl-CoA synthetase long-chain family member 3 |
| chr6_-_67353131 | 1.01 |
ENSMUST00000018485.4
|
Il12rb2
|
interleukin 12 receptor, beta 2 |
| chr2_-_30249202 | 1.01 |
ENSMUST00000100215.11
ENSMUST00000113620.10 ENSMUST00000163668.3 ENSMUST00000028214.15 ENSMUST00000113621.10 |
Sh3glb2
|
SH3-domain GRB2-like endophilin B2 |
| chr1_-_173105344 | 1.01 |
ENSMUST00000111224.5
|
Mptx2
|
mucosal pentraxin 2 |
| chr1_-_175319842 | 1.00 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr6_+_79995994 | 0.99 |
ENSMUST00000126399.2
ENSMUST00000136421.2 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr7_+_140808680 | 0.98 |
ENSMUST00000106027.9
|
Phrf1
|
PHD and ring finger domains 1 |
| chr6_+_68916540 | 0.98 |
ENSMUST00000103339.2
|
Igkv13-84
|
immunoglobulin kappa chain variable 13-84 |
| chr6_-_69609162 | 0.98 |
ENSMUST00000199437.2
|
Igkv4-54
|
immunoglobulin kappa chain variable 4-54 |
| chr12_-_99849660 | 0.96 |
ENSMUST00000221929.2
ENSMUST00000046485.5 |
Efcab11
|
EF-hand calcium binding domain 11 |
| chr17_-_35381945 | 0.96 |
ENSMUST00000174805.2
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr17_-_30790804 | 0.96 |
ENSMUST00000236799.2
ENSMUST00000237048.2 |
Btbd9
|
BTB (POZ) domain containing 9 |
| chrX_+_113384008 | 0.96 |
ENSMUST00000113371.8
ENSMUST00000040504.12 |
Klhl4
|
kelch-like 4 |
| chr11_+_78816002 | 0.96 |
ENSMUST00000214397.3
|
Nos2
|
nitric oxide synthase 2, inducible |
| chr11_-_11920540 | 0.95 |
ENSMUST00000109653.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chr17_+_33212143 | 0.95 |
ENSMUST00000087666.11
ENSMUST00000157017.2 |
Zfp952
|
zinc finger protein 952 |
| chr16_-_57427179 | 0.95 |
ENSMUST00000114371.5
ENSMUST00000232413.2 |
Cmss1
|
cms small ribosomal subunit 1 |
| chr7_+_6998299 | 0.94 |
ENSMUST00000208049.2
ENSMUST00000086248.7 ENSMUST00000208518.2 ENSMUST00000207711.2 |
Aurkc
|
aurora kinase C |
| chr15_-_83949528 | 0.93 |
ENSMUST00000156187.8
|
Efcab6
|
EF-hand calcium binding domain 6 |
| chr7_-_79570342 | 0.93 |
ENSMUST00000075657.8
|
Ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chrX_+_136552469 | 0.92 |
ENSMUST00000075471.4
|
Il1rapl2
|
interleukin 1 receptor accessory protein-like 2 |
| chrX_-_149879501 | 0.92 |
ENSMUST00000112683.9
ENSMUST00000026295.10 |
Tsr2
|
TSR2 20S rRNA accumulation |
| chr14_-_47514248 | 0.91 |
ENSMUST00000187531.8
ENSMUST00000111790.2 |
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr8_+_21777425 | 0.91 |
ENSMUST00000098893.4
|
Defa3
|
defensin, alpha, 3 |
| chr18_+_69726431 | 0.90 |
ENSMUST00000201091.4
ENSMUST00000201037.4 ENSMUST00000114977.5 |
Tcf4
|
transcription factor 4 |
| chr11_+_109540201 | 0.90 |
ENSMUST00000106677.8
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr13_+_19524136 | 0.90 |
ENSMUST00000103564.3
|
Trgv1
|
T cell receptor gamma, variable 1 |
| chrX_-_59193003 | 0.89 |
ENSMUST00000033478.5
ENSMUST00000101531.10 |
Mcf2
|
mcf.2 transforming sequence |
| chr13_+_69950509 | 0.89 |
ENSMUST00000223376.2
ENSMUST00000222387.2 |
Med10
|
mediator complex subunit 10 |
| chr12_+_17398421 | 0.89 |
ENSMUST00000046011.12
|
Nol10
|
nucleolar protein 10 |
| chrY_-_1245685 | 0.88 |
ENSMUST00000143286.8
ENSMUST00000137048.8 ENSMUST00000069309.14 ENSMUST00000139365.8 ENSMUST00000154004.8 |
Uty
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr14_+_14986011 | 0.88 |
ENSMUST00000164366.8
|
Gm3752
|
predicted gene 3752 |
| chr1_+_132405099 | 0.87 |
ENSMUST00000190825.7
ENSMUST00000190997.7 ENSMUST00000187505.7 ENSMUST00000027700.15 ENSMUST00000188575.7 |
Rbbp5
|
retinoblastoma binding protein 5, histone lysine methyltransferase complex subunit |
| chr6_-_136918844 | 0.85 |
ENSMUST00000204934.2
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr16_+_49620883 | 0.85 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chrX_-_69408627 | 0.84 |
ENSMUST00000101509.9
|
Ids
|
iduronate 2-sulfatase |
| chr16_+_48877762 | 0.84 |
ENSMUST00000168680.2
|
Myh15
|
myosin, heavy chain 15 |
| chr10_+_85222677 | 0.84 |
ENSMUST00000105307.8
ENSMUST00000020231.10 |
Btbd11
|
BTB (POZ) domain containing 11 |
| chr8_-_86091946 | 0.84 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr1_-_160134873 | 0.84 |
ENSMUST00000193185.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr2_-_121304521 | 0.83 |
ENSMUST00000056732.4
|
Mfap1b
|
microfibrillar-associated protein 1B |
| chr17_+_33212097 | 0.81 |
ENSMUST00000141815.3
|
Zfp952
|
zinc finger protein 952 |
| chr4_+_103000248 | 0.81 |
ENSMUST00000106855.2
|
Mier1
|
MEIR1 treanscription regulator |
| chr3_-_129834788 | 0.81 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr15_+_79543397 | 0.81 |
ENSMUST00000023064.9
|
Cby1
|
chibby family member 1, beta catenin antagonist |
| chr12_-_40298072 | 0.80 |
ENSMUST00000169926.8
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr3_-_27950491 | 0.80 |
ENSMUST00000058077.4
|
Tmem212
|
transmembrane protein 212 |
| chr16_+_44167484 | 0.80 |
ENSMUST00000050897.7
|
Spice1
|
spindle and centriole associated protein 1 |
| chr9_+_74959259 | 0.79 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr11_-_58794999 | 0.79 |
ENSMUST00000102703.2
|
Zfp39
|
zinc finger protein 39 |
| chr6_+_85564506 | 0.78 |
ENSMUST00000072018.6
|
Alms1
|
ALMS1, centrosome and basal body associated |
| chr12_+_65122355 | 0.78 |
ENSMUST00000058889.5
|
Fancm
|
Fanconi anemia, complementation group M |
| chr6_+_29398919 | 0.78 |
ENSMUST00000181464.8
ENSMUST00000180829.8 |
Ccdc136
|
coiled-coil domain containing 136 |
| chr5_+_134611544 | 0.78 |
ENSMUST00000023867.8
|
Rfc2
|
replication factor C (activator 1) 2 |
| chr8_+_36561982 | 0.77 |
ENSMUST00000110492.2
|
Prag1
|
PEAK1 related kinase activating pseudokinase 1 |
| chr10_+_24471340 | 0.77 |
ENSMUST00000020171.12
|
Ccn2
|
cellular communication network factor 2 |
| chr6_-_69877642 | 0.77 |
ENSMUST00000103370.3
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr7_+_101750943 | 0.76 |
ENSMUST00000033300.4
|
Art1
|
ADP-ribosyltransferase 1 |
| chr2_-_180928867 | 0.76 |
ENSMUST00000130475.8
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr12_-_110807330 | 0.76 |
ENSMUST00000177224.2
ENSMUST00000084974.11 ENSMUST00000070565.15 |
Mok
|
MOK protein kinase |
| chr1_+_78635591 | 0.75 |
ENSMUST00000134566.8
ENSMUST00000142704.8 ENSMUST00000053760.12 |
Acsl3
Utp14b
|
acyl-CoA synthetase long-chain family member 3 UTP14B small subunit processome component |
| chr16_-_16962279 | 0.75 |
ENSMUST00000232033.2
ENSMUST00000231597.2 ENSMUST00000232540.2 |
Ccdc116
|
coiled-coil domain containing 116 |
| chr1_+_121358778 | 0.74 |
ENSMUST00000036025.16
ENSMUST00000112621.2 |
Ccdc93
|
coiled-coil domain containing 93 |
| chr2_-_126342551 | 0.74 |
ENSMUST00000129187.2
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chrX_+_52001108 | 0.74 |
ENSMUST00000078944.13
ENSMUST00000101587.10 ENSMUST00000154864.4 |
Phf6
|
PHD finger protein 6 |
| chr7_+_35096470 | 0.73 |
ENSMUST00000079414.12
|
Cep89
|
centrosomal protein 89 |
| chr6_-_39396691 | 0.73 |
ENSMUST00000146785.8
ENSMUST00000114823.8 |
Mkrn1
|
makorin, ring finger protein, 1 |
| chr11_-_102907991 | 0.72 |
ENSMUST00000021313.9
|
Dcakd
|
dephospho-CoA kinase domain containing |
| chr17_+_31220910 | 0.71 |
ENSMUST00000235827.2
|
Umodl1
|
uromodulin-like 1 |
| chr11_-_94133527 | 0.70 |
ENSMUST00000061469.4
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr6_+_89691185 | 0.70 |
ENSMUST00000075158.2
|
Vmn1r40
|
vomeronasal 1 receptor 40 |
| chr1_+_59724108 | 0.69 |
ENSMUST00000027174.10
ENSMUST00000190231.7 ENSMUST00000191142.7 ENSMUST00000185772.7 |
Nop58
|
NOP58 ribonucleoprotein |
| chr18_-_24663260 | 0.69 |
ENSMUST00000046206.5
|
Rprd1a
|
regulation of nuclear pre-mRNA domain containing 1A |
| chr13_-_19579961 | 0.68 |
ENSMUST00000039694.13
|
Stard3nl
|
STARD3 N-terminal like |
| chr15_+_5215000 | 0.67 |
ENSMUST00000118193.8
ENSMUST00000022751.15 |
Ttc33
|
tetratricopeptide repeat domain 33 |
| chr7_+_19016536 | 0.67 |
ENSMUST00000032559.17
|
Rtn2
|
reticulon 2 (Z-band associated protein) |
| chr2_+_69869459 | 0.67 |
ENSMUST00000060208.11
|
Myo3b
|
myosin IIIB |
| chr4_+_132366298 | 0.67 |
ENSMUST00000135299.8
ENSMUST00000020197.14 ENSMUST00000180250.8 ENSMUST00000081726.13 ENSMUST00000079157.11 |
Eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr9_-_20864096 | 0.67 |
ENSMUST00000004202.17
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr9_-_36637923 | 0.67 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
| chr17_+_17594808 | 0.66 |
ENSMUST00000232396.2
ENSMUST00000232199.2 |
Riok2
|
RIO kinase 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.7 | 5.9 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.6 | 2.5 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.6 | 2.4 | GO:0072218 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.5 | 3.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.5 | 1.5 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.5 | 1.4 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.4 | 1.8 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.4 | 1.6 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.4 | 1.6 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) |
| 0.4 | 2.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 1.5 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.4 | 1.5 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.4 | 1.1 | GO:0002477 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.4 | 1.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 | 1.9 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.3 | 1.5 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.3 | 1.3 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.2 | 0.2 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.2 | 1.9 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.2 | 0.7 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.2 | 1.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 1.9 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.2 | 1.4 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 2.4 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.2 | 1.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 1.6 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 1.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 0.9 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.2 | 1.5 | GO:0009313 | ganglioside catabolic process(GO:0006689) penetration of zona pellucida(GO:0007341) oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 1.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.2 | 1.1 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.2 | 2.1 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 | 1.9 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.2 | 1.2 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.2 | 0.7 | GO:0090309 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 0.7 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.2 | 0.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 2.6 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.2 | 1.3 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 1.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 1.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 2.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 1.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.8 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.5 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.1 | 2.3 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 0.1 | 1.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.1 | 0.9 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 1.8 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 2.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.5 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 0.7 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.4 | GO:0002590 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.1 | 1.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.4 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.9 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 1.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 1.1 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 1.0 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 0.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 1.0 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.1 | 0.6 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.1 | 1.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.2 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.1 | 0.4 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.1 | 0.3 | GO:0000389 | generation of catalytic spliceosome for second transesterification step(GO:0000350) mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.9 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.1 | 0.3 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.8 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.7 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.0 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 3.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 2.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 2.0 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.1 | 0.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.6 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.6 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.1 | 0.7 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 1.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.7 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 1.5 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 1.0 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.5 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.1 | 0.7 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 1.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.5 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.7 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.7 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.5 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.6 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 1.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.2 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 9.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.8 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 1.2 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.8 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.8 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 1.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.1 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.3 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.3 | GO:0006348 | DNA replication-dependent nucleosome assembly(GO:0006335) chromatin silencing at telomere(GO:0006348) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.5 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 1.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.8 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 1.1 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 3.2 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 1.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.3 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.8 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.4 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 1.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 3.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.8 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 1.1 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 1.7 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0032976 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 0.0 | 0.9 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.5 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 0.4 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 | 0.6 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.0 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 | 0.2 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 1.0 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 1.6 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.7 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.8 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 1.6 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.7 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.0 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.2 | 3.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.8 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.2 | 1.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 0.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 5.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 0.7 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.2 | 1.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 0.5 | GO:0005712 | chiasma(GO:0005712) late recombination nodule(GO:0005715) |
| 0.2 | 0.9 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 3.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 2.0 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 1.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.6 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.5 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.8 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 2.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.2 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 1.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 2.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 1.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.8 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 1.4 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.7 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 1.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.0 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 2.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 2.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 3.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 2.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.6 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0097144 | BAX complex(GO:0097144) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 7.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 4.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 2.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 2.9 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.9 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.5 | 3.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.5 | 1.4 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.5 | 2.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.4 | 1.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.4 | 1.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.3 | 1.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 1.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.3 | 1.5 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.2 | 0.7 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.2 | 1.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.2 | 3.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 0.9 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 1.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 1.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 2.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 1.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 1.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 1.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 0.3 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.2 | 0.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.2 | 0.5 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.2 | 0.5 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.2 | 1.3 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.5 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.2 | 4.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.9 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 1.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 1.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.9 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.5 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 0.4 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.6 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.4 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 2.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.4 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 2.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 2.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 2.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.2 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 0.8 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.3 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 1.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 1.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.9 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 3.8 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 2.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 2.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.6 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 1.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 1.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 1.0 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 1.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 2.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 1.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 3.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.4 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 3.4 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 1.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 2.4 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 2.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 1.6 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 3.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 2.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 3.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 5.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.7 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 5.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 2.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 3.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 2.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.8 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 2.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 4.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |