Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
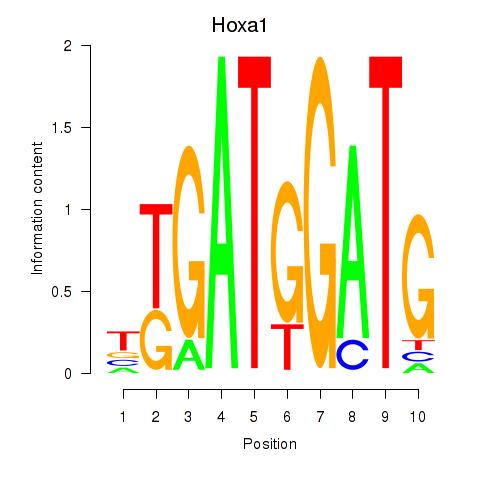
Results for Hoxa1
Z-value: 1.10
Transcription factors associated with Hoxa1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa1
|
ENSMUSG00000029844.10 | Hoxa1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa1 | mm39_v1_chr6_-_52135261_52135332 | 0.07 | 5.8e-01 | Click! |
Activity profile of Hoxa1 motif
Sorted Z-values of Hoxa1 motif
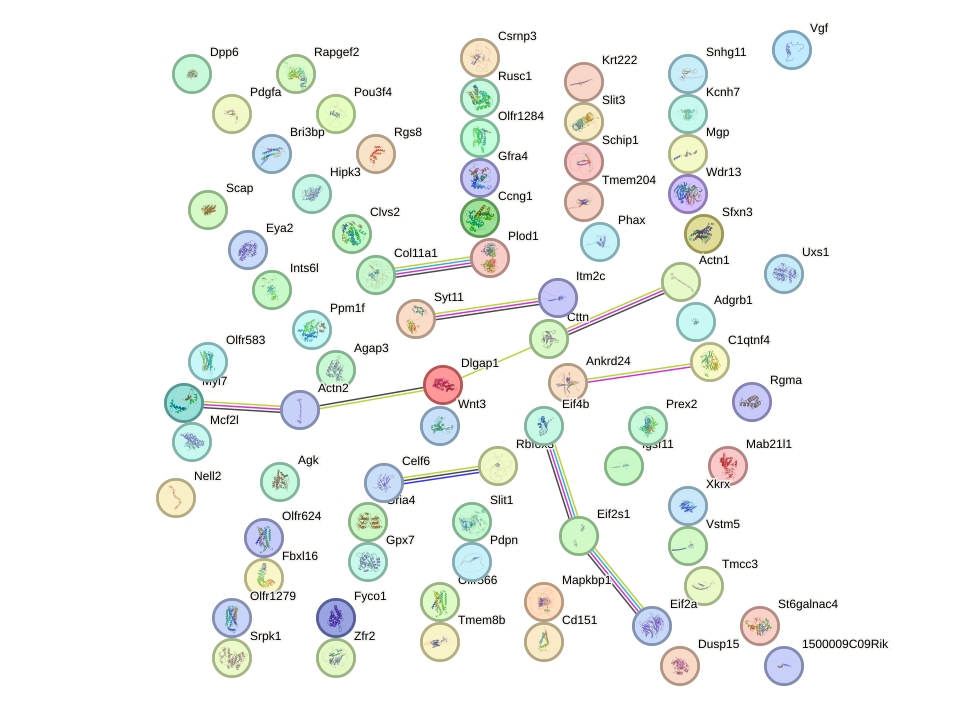
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_158217558 | 22.06 |
ENSMUST00000109488.8
|
Snhg11
|
small nucleolar RNA host gene 11 |
| chr15_-_95426419 | 14.25 |
ENSMUST00000229933.2
ENSMUST00000166170.9 |
Nell2
|
NEL-like 2 |
| chr4_-_143026068 | 11.90 |
ENSMUST00000030317.14
|
Pdpn
|
podoplanin |
| chr15_+_82136598 | 11.44 |
ENSMUST00000136948.3
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr13_-_12355604 | 10.18 |
ENSMUST00000168193.8
ENSMUST00000064204.14 |
Actn2
|
actinin alpha 2 |
| chr10_+_81464370 | 9.31 |
ENSMUST00000123896.8
ENSMUST00000153573.2 ENSMUST00000119336.8 |
Ankrd24
|
ankyrin repeat domain 24 |
| chr11_-_99134885 | 7.91 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr17_+_70829050 | 7.51 |
ENSMUST00000133717.9
ENSMUST00000148486.8 |
Dlgap1
|
DLG associated protein 1 |
| chr3_-_88669551 | 7.39 |
ENSMUST00000183267.2
|
Syt11
|
synaptotagmin XI |
| chr17_+_26036893 | 7.27 |
ENSMUST00000235694.2
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr14_+_66581818 | 6.70 |
ENSMUST00000118426.8
ENSMUST00000121955.8 ENSMUST00000120229.8 ENSMUST00000134440.2 |
Stmn4
|
stathmin-like 4 |
| chr10_-_33500583 | 6.52 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr17_+_70829144 | 6.38 |
ENSMUST00000140728.8
|
Dlgap1
|
DLG associated protein 1 |
| chr7_+_73040908 | 6.31 |
ENSMUST00000128471.2
ENSMUST00000139780.3 |
Rgma
|
repulsive guidance molecule family member A |
| chr14_+_70314727 | 6.31 |
ENSMUST00000225200.2
|
Egr3
|
early growth response 3 |
| chr5_+_27466914 | 6.30 |
ENSMUST00000101471.4
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr10_+_81068980 | 6.30 |
ENSMUST00000144087.2
ENSMUST00000117798.8 |
Zfr2
|
zinc finger RNA binding protein 2 |
| chr14_+_66581745 | 6.28 |
ENSMUST00000152093.8
ENSMUST00000074523.13 |
Stmn4
|
stathmin-like 4 |
| chr17_-_90763300 | 6.19 |
ENSMUST00000159778.8
ENSMUST00000174337.8 ENSMUST00000172466.8 |
Nrxn1
|
neurexin I |
| chr14_+_70314652 | 5.56 |
ENSMUST00000035908.3
|
Egr3
|
early growth response 3 |
| chr2_-_130885008 | 5.54 |
ENSMUST00000110240.10
ENSMUST00000066958.11 ENSMUST00000110235.2 |
Gfra4
|
glial cell line derived neurotrophic factor family receptor alpha 4 |
| chr9_+_15150341 | 5.37 |
ENSMUST00000034413.8
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr15_+_74388044 | 5.29 |
ENSMUST00000042035.16
|
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr5_+_137059127 | 5.24 |
ENSMUST00000041543.9
ENSMUST00000186451.2 |
Vgf
|
VGF nerve growth factor inducible |
| chr2_+_165436929 | 5.11 |
ENSMUST00000088132.13
|
Eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr14_-_104081827 | 5.10 |
ENSMUST00000022718.11
|
Ednrb
|
endothelin receptor type B |
| chr8_+_12999480 | 5.05 |
ENSMUST00000110866.9
|
Mcf2l
|
mcf.2 transforming sequence-like |
| chr8_+_12999394 | 4.86 |
ENSMUST00000110867.9
|
Mcf2l
|
mcf.2 transforming sequence-like |
| chr5_+_24679154 | 4.79 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr2_-_130884602 | 4.72 |
ENSMUST00000028787.12
ENSMUST00000110239.8 ENSMUST00000110234.8 |
Gfra4
|
glial cell line derived neurotrophic factor family receptor alpha 4 |
| chr2_+_65676111 | 4.71 |
ENSMUST00000122912.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr9_-_4796217 | 4.71 |
ENSMUST00000027020.13
ENSMUST00000163309.2 |
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
| chr4_+_101365144 | 4.56 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr19_+_45036037 | 4.54 |
ENSMUST00000062213.13
|
Sfxn3
|
sideroflexin 3 |
| chr19_+_45036220 | 4.45 |
ENSMUST00000084493.8
|
Sfxn3
|
sideroflexin 3 |
| chr2_+_90716204 | 4.44 |
ENSMUST00000111466.3
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4 |
| chr2_+_156317416 | 4.44 |
ENSMUST00000029155.16
|
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr19_+_45035942 | 4.26 |
ENSMUST00000237222.2
ENSMUST00000111954.11 |
Sfxn3
|
sideroflexin 3 |
| chr3_+_68479578 | 4.18 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr4_-_106656976 | 4.00 |
ENSMUST00000145061.8
ENSMUST00000102762.10 |
Acot11
|
acyl-CoA thioesterase 11 |
| chr5_+_88731366 | 3.94 |
ENSMUST00000199312.5
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr4_+_101365052 | 3.93 |
ENSMUST00000038207.12
|
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr5_+_88731386 | 3.85 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr1_+_11063678 | 3.68 |
ENSMUST00000027056.12
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr1_+_153541020 | 3.64 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr4_-_143026033 | 3.61 |
ENSMUST00000119654.2
|
Pdpn
|
podoplanin |
| chr9_-_4796142 | 3.60 |
ENSMUST00000063508.15
|
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
| chr2_+_65676176 | 3.55 |
ENSMUST00000053910.10
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr3_-_89000591 | 3.43 |
ENSMUST00000090929.12
ENSMUST00000052539.13 |
Rusc1
|
RUN and SH3 domain containing 1 |
| chr10_+_94412116 | 3.41 |
ENSMUST00000117929.2
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr4_+_43669266 | 3.29 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr19_-_41732104 | 2.83 |
ENSMUST00000025993.10
|
Slit1
|
slit guidance ligand 1 |
| chr5_+_137059522 | 2.79 |
ENSMUST00000187382.2
|
Vgf
|
VGF nerve growth factor inducible |
| chr1_+_85822250 | 2.66 |
ENSMUST00000185569.2
|
Itm2c
|
integral membrane protein 2C |
| chr6_-_136852792 | 2.59 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chrX_+_55500170 | 2.51 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr11_-_118460736 | 2.49 |
ENSMUST00000136551.3
|
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr6_+_40302106 | 2.44 |
ENSMUST00000031977.12
|
Agk
|
acylglycerol kinase |
| chr17_-_25300112 | 2.28 |
ENSMUST00000024984.7
|
Tmem204
|
transmembrane protein 204 |
| chr17_-_28841006 | 2.10 |
ENSMUST00000233353.2
|
Srpk1
|
serine/arginine-rich protein specific kinase 1 |
| chr3_-_79053182 | 2.09 |
ENSMUST00000118340.7
|
Rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr2_+_119803230 | 2.05 |
ENSMUST00000229024.2
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr3_-_57202546 | 2.02 |
ENSMUST00000196506.2
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chrX_+_109857866 | 1.97 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr2_+_32477069 | 1.93 |
ENSMUST00000102818.11
|
St6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr4_-_106657446 | 1.90 |
ENSMUST00000148688.2
|
Acot11
|
acyl-CoA thioesterase 11 |
| chr8_-_91544021 | 1.79 |
ENSMUST00000209208.2
|
Gm19935
|
predicted gene, 19935 |
| chr9_+_121727421 | 1.75 |
ENSMUST00000214340.2
ENSMUST00000050327.5 |
Ackr2
|
atypical chemokine receptor 2 |
| chr3_+_113824181 | 1.62 |
ENSMUST00000123619.8
ENSMUST00000092155.12 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr17_-_28841370 | 1.62 |
ENSMUST00000233129.2
ENSMUST00000232751.2 ENSMUST00000233794.2 ENSMUST00000114767.3 |
Srpk1
|
serine/arginine-rich protein specific kinase 1 |
| chr7_-_144024451 | 1.59 |
ENSMUST00000033407.13
|
Cttn
|
cortactin |
| chr16_+_16714333 | 1.49 |
ENSMUST00000027373.12
ENSMUST00000232247.2 |
Ppm1f
|
protein phosphatase 1F (PP2C domain containing) |
| chr16_+_38722666 | 1.40 |
ENSMUST00000023478.8
|
Igsf11
|
immunoglobulin superfamily, member 11 |
| chr16_-_10360893 | 1.39 |
ENSMUST00000184863.8
ENSMUST00000038281.6 |
Dexi
|
dexamethasone-induced transcript |
| chr11_+_24030663 | 1.33 |
ENSMUST00000118955.2
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr4_-_108263873 | 1.32 |
ENSMUST00000184609.2
|
Gpx7
|
glutathione peroxidase 7 |
| chr7_-_102507962 | 1.30 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr9_+_110173253 | 1.27 |
ENSMUST00000199709.3
|
Scap
|
SREBF chaperone |
| chr2_+_111136546 | 1.25 |
ENSMUST00000090329.2
|
Olfr1279
|
olfactory receptor 1279 |
| chr15_+_89095724 | 1.20 |
ENSMUST00000227951.2
ENSMUST00000226221.2 ENSMUST00000238818.2 ENSMUST00000228284.2 |
Ppp6r2
|
protein phosphatase 6, regulatory subunit 2 |
| chrX_-_7999009 | 1.15 |
ENSMUST00000130832.8
ENSMUST00000033506.13 ENSMUST00000115623.8 ENSMUST00000153839.2 |
Wdr13
|
WD repeat domain 13 |
| chr12_-_80306865 | 1.11 |
ENSMUST00000167327.2
|
Actn1
|
actinin, alpha 1 |
| chr1_-_43866910 | 1.10 |
ENSMUST00000153317.6
ENSMUST00000128261.2 ENSMUST00000126008.8 ENSMUST00000139451.8 |
Uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr11_-_40646090 | 1.06 |
ENSMUST00000020576.8
|
Ccng1
|
cyclin G1 |
| chr3_-_152931392 | 0.98 |
ENSMUST00000199707.2
|
St6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr2_-_63014622 | 0.87 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr2_+_119803180 | 0.81 |
ENSMUST00000066058.8
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr15_+_9140614 | 0.81 |
ENSMUST00000227556.3
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr11_-_5848771 | 0.81 |
ENSMUST00000102921.4
|
Myl7
|
myosin, light polypeptide 7, regulatory |
| chr5_+_125518609 | 0.79 |
ENSMUST00000049040.14
|
Bri3bp
|
Bri3 binding protein |
| chr16_+_22926162 | 0.78 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr18_+_56695515 | 0.71 |
ENSMUST00000130163.8
ENSMUST00000132628.8 |
Phax
|
phosphorylated adaptor for RNA export |
| chr7_-_103320398 | 0.71 |
ENSMUST00000062144.4
|
Olfr624
|
olfactory receptor 624 |
| chr4_+_128582519 | 0.66 |
ENSMUST00000106080.8
|
Phc2
|
polyhomeotic 2 |
| chr3_+_58433236 | 0.58 |
ENSMUST00000029387.15
|
Eif2a
|
eukaryotic translation initiation factor 2A |
| chr9_+_58036345 | 0.56 |
ENSMUST00000085677.9
|
Stra6
|
stimulated by retinoic acid gene 6 |
| chr7_+_141048191 | 0.54 |
ENSMUST00000211071.2
|
Cd151
|
CD151 antigen |
| chr9_+_59496571 | 0.54 |
ENSMUST00000121266.8
ENSMUST00000118164.3 |
Celf6
|
CUGBP, Elav-like family member 6 |
| chr2_-_152793469 | 0.50 |
ENSMUST00000037715.7
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr10_+_73782857 | 0.49 |
ENSMUST00000191709.6
ENSMUST00000193739.6 ENSMUST00000195531.6 |
Pcdh15
|
protocadherin 15 |
| chr3_+_55689921 | 0.48 |
ENSMUST00000075422.6
|
Mab21l1
|
mab-21-like 1 |
| chr3_+_58913234 | 0.48 |
ENSMUST00000040846.15
|
Med12l
|
mediator complex subunit 12-like |
| chr7_+_106513407 | 0.44 |
ENSMUST00000098140.2
|
Olfr1532-ps1
|
olfactory receptor 1532, pseudogene 1 |
| chr2_-_104324035 | 0.42 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr4_-_35845204 | 0.41 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr4_-_148021159 | 0.34 |
ENSMUST00000105712.2
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr5_-_138981842 | 0.33 |
ENSMUST00000110897.8
|
Pdgfa
|
platelet derived growth factor, alpha |
| chrX_-_133062677 | 0.32 |
ENSMUST00000033611.5
|
Xkrx
|
X-linked Kx blood group related, X-linked |
| chr5_+_43672856 | 0.28 |
ENSMUST00000076939.10
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr11_-_102187445 | 0.28 |
ENSMUST00000107132.3
ENSMUST00000073234.9 |
Atxn7l3
|
ataxin 7-like 3 |
| chr11_+_103664976 | 0.27 |
ENSMUST00000000127.6
|
Wnt3
|
wingless-type MMTV integration site family, member 3 |
| chr18_-_56695333 | 0.24 |
ENSMUST00000066208.13
ENSMUST00000172734.8 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr11_+_4833186 | 0.22 |
ENSMUST00000139737.2
|
Nipsnap1
|
nipsnap homolog 1 |
| chr16_-_18695267 | 0.20 |
ENSMUST00000119273.2
|
Mrpl40
|
mitochondrial ribosomal protein L40 |
| chr2_-_165717697 | 0.18 |
ENSMUST00000153655.8
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr13_+_113429690 | 0.15 |
ENSMUST00000136755.10
|
Cspg4b
|
chondroitin sulfate proteoglycan 4B |
| chr1_+_74700952 | 0.10 |
ENSMUST00000129890.8
|
Ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr3_-_126918491 | 0.08 |
ENSMUST00000238781.2
|
Ank2
|
ankyrin 2, brain |
| chr1_-_157240138 | 0.07 |
ENSMUST00000078308.13
|
Rasal2
|
RAS protein activator like 2 |
| chr7_+_103628383 | 0.06 |
ENSMUST00000098185.2
|
Olfr635
|
olfactory receptor 635 |
| chr9_+_58036604 | 0.00 |
ENSMUST00000034880.10
|
Stra6
|
stimulated by retinoic acid gene 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 3.4 | 10.2 | GO:0086097 | actin filament uncapping(GO:0051695) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 1.8 | 7.4 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 1.4 | 8.5 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 1.1 | 13.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.0 | 2.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 1.0 | 6.2 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.9 | 5.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.8 | 14.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.8 | 11.9 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.5 | 8.0 | GO:0043084 | penile erection(GO:0043084) |
| 0.5 | 2.8 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.4 | 6.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.4 | 4.8 | GO:0030578 | PML body organization(GO:0030578) |
| 0.4 | 2.9 | GO:0007256 | activation of JNKK activity(GO:0007256) negative regulation of defense response to bacterium(GO:1900425) |
| 0.3 | 2.7 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 1.3 | GO:1904799 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.3 | 6.3 | GO:1900121 | negative regulation of axon regeneration(GO:0048681) negative regulation of receptor binding(GO:1900121) |
| 0.2 | 7.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 3.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 0.7 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.2 | 4.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 9.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 8.3 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.2 | 13.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.5 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 0.6 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 1.6 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.1 | 0.3 | GO:0035789 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.1 | 2.0 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 6.4 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.1 | 1.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.3 | GO:1904954 | Spemann organizer formation(GO:0060061) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.1 | 4.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 6.5 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 10.3 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 3.7 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 8.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.6 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 2.3 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 7.4 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.1 | 3.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.6 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.1 | 0.3 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 1.3 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 5.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 2.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 2.4 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 1.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 2.6 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 4.4 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.2 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 1.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 3.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 15.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.0 | 8.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.5 | 7.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.3 | 7.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.3 | 6.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 10.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 0.6 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 1.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 2.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 3.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 16.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 8.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 7.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 16.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 33.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 0.6 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 7.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 6.5 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 9.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 5.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 6.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 2.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.8 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 3.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 4.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 8.4 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 13.1 | GO:0043005 | neuron projection(GO:0043005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 2.0 | 10.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 1.7 | 5.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 1.0 | 8.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.8 | 2.4 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.7 | 15.5 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.5 | 2.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.5 | 6.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.4 | 6.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.3 | 2.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.3 | 9.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 2.9 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 8.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 5.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 6.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 14.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 7.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 3.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 13.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 2.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 6.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 4.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 3.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 4.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 13.1 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 8.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 3.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 13.8 | GO:0019904 | protein domain specific binding(GO:0019904) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 11.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 13.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 6.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 19.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 5.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 3.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 3.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 4.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 18.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 6.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 4.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 9.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 6.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 3.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 4.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |