Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
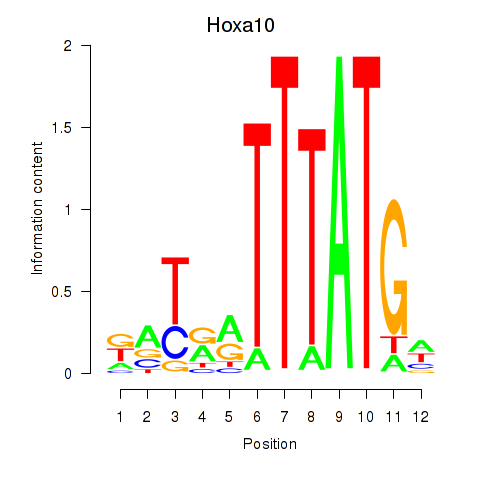
Results for Hoxa10
Z-value: 0.86
Transcription factors associated with Hoxa10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa10
|
ENSMUSG00000000938.12 | Hoxa10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa10 | mm39_v1_chr6_-_52211882_52211940 | -0.41 | 3.7e-04 | Click! |
Activity profile of Hoxa10 motif
Sorted Z-values of Hoxa10 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa10
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_110781268 | 7.27 |
ENSMUST00000099623.10
|
Ano3
|
anoctamin 3 |
| chr2_+_102488985 | 6.91 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_+_83642910 | 6.04 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chrX_+_158242121 | 6.03 |
ENSMUST00000112470.3
ENSMUST00000043151.12 ENSMUST00000156172.3 |
Map7d2
|
MAP7 domain containing 2 |
| chr1_+_66360865 | 6.02 |
ENSMUST00000114013.8
|
Map2
|
microtubule-associated protein 2 |
| chr5_-_118382926 | 5.18 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr6_+_47021979 | 5.14 |
ENSMUST00000150737.2
|
Cntnap2
|
contactin associated protein-like 2 |
| chr3_+_67799510 | 5.06 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr14_+_27598021 | 4.98 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr14_-_9015639 | 4.89 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chr16_+_41353360 | 4.76 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr2_-_79738773 | 4.63 |
ENSMUST00000102652.10
ENSMUST00000102651.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_-_70999547 | 4.47 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chr8_-_34237752 | 4.46 |
ENSMUST00000179364.3
|
Smim18
|
small integral membrane protein 18 |
| chr3_-_59170245 | 4.31 |
ENSMUST00000050360.14
ENSMUST00000199609.2 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr7_-_45019984 | 4.28 |
ENSMUST00000003971.10
|
Lin7b
|
lin-7 homolog B (C. elegans) |
| chr5_-_103247920 | 4.11 |
ENSMUST00000112848.8
|
Mapk10
|
mitogen-activated protein kinase 10 |
| chr2_-_79738734 | 4.09 |
ENSMUST00000090756.11
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr11_-_99313078 | 3.94 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr19_+_43428843 | 3.82 |
ENSMUST00000223787.2
ENSMUST00000165311.3 |
Cnnm1
|
cyclin M1 |
| chr3_-_94343874 | 3.75 |
ENSMUST00000204913.3
ENSMUST00000191506.8 ENSMUST00000199678.4 |
Oaz3
|
ornithine decarboxylase antizyme 3 |
| chr9_+_21848282 | 3.52 |
ENSMUST00000046371.13
|
Plppr2
|
phospholipid phosphatase related 2 |
| chr14_-_9015757 | 3.42 |
ENSMUST00000153954.8
|
Synpr
|
synaptoporin |
| chr4_+_41966058 | 3.41 |
ENSMUST00000108026.3
|
Fam205a4
|
family with sequence similarity 205, member A4 |
| chr13_-_99653045 | 3.39 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr18_-_12993257 | 3.31 |
ENSMUST00000124570.3
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr12_-_72964646 | 3.30 |
ENSMUST00000044000.12
|
4930447C04Rik
|
RIKEN cDNA 4930447C04 gene |
| chr11_+_42310557 | 3.28 |
ENSMUST00000007797.10
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr8_+_59365291 | 3.06 |
ENSMUST00000160055.2
|
BC030500
|
cDNA sequence BC030500 |
| chr17_+_17669082 | 3.00 |
ENSMUST00000140134.2
|
Lix1
|
limb and CNS expressed 1 |
| chr6_-_42686970 | 2.93 |
ENSMUST00000045054.11
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr11_+_87473027 | 2.93 |
ENSMUST00000133202.3
|
Septin4
|
septin 4 |
| chr1_-_125840838 | 2.93 |
ENSMUST00000161361.3
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr11_-_41891111 | 2.86 |
ENSMUST00000109290.2
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr1_-_124773767 | 2.83 |
ENSMUST00000239072.2
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr10_-_10958031 | 2.80 |
ENSMUST00000105561.9
ENSMUST00000044306.13 |
Grm1
|
glutamate receptor, metabotropic 1 |
| chr4_-_119240885 | 2.50 |
ENSMUST00000238422.2
ENSMUST00000238719.2 ENSMUST00000238723.2 ENSMUST00000044781.9 ENSMUST00000084307.5 ENSMUST00000148236.9 ENSMUST00000127474.3 ENSMUST00000238704.2 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr4_-_94817025 | 2.46 |
ENSMUST00000030309.6
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr4_-_138053545 | 2.44 |
ENSMUST00000105817.4
|
Pink1
|
PTEN induced putative kinase 1 |
| chr12_-_64521464 | 2.43 |
ENSMUST00000059833.8
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr9_+_109865810 | 2.35 |
ENSMUST00000163190.8
|
Map4
|
microtubule-associated protein 4 |
| chr8_+_13034245 | 2.35 |
ENSMUST00000110873.10
ENSMUST00000173006.8 ENSMUST00000145067.8 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr13_+_83652352 | 2.33 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr4_-_102883905 | 2.29 |
ENSMUST00000084382.6
ENSMUST00000106869.3 |
Insl5
|
insulin-like 5 |
| chr8_+_94537910 | 2.29 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr17_-_6129968 | 2.25 |
ENSMUST00000024570.6
ENSMUST00000097432.10 |
Serac1
|
serine active site containing 1 |
| chr1_-_73055043 | 2.25 |
ENSMUST00000027374.7
|
Tnp1
|
transition protein 1 |
| chr13_+_83720484 | 2.24 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr11_-_107238956 | 2.23 |
ENSMUST00000134763.2
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr9_-_112061517 | 2.19 |
ENSMUST00000035085.12
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr4_-_94817056 | 2.19 |
ENSMUST00000107097.9
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr4_-_119241024 | 2.18 |
ENSMUST00000127149.8
ENSMUST00000152879.9 ENSMUST00000238673.2 ENSMUST00000238485.2 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr18_+_37840092 | 2.06 |
ENSMUST00000195823.2
|
Pcdhga6
|
protocadherin gamma subfamily A, 6 |
| chr5_+_27109679 | 2.04 |
ENSMUST00000120555.8
|
Dpp6
|
dipeptidylpeptidase 6 |
| chrX_-_42363663 | 2.04 |
ENSMUST00000016294.8
|
Tenm1
|
teneurin transmembrane protein 1 |
| chr17_+_6025861 | 2.03 |
ENSMUST00000142409.8
ENSMUST00000061091.14 |
Synj2
|
synaptojanin 2 |
| chr17_+_6026015 | 2.00 |
ENSMUST00000115790.8
|
Synj2
|
synaptojanin 2 |
| chr15_+_21111428 | 1.99 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr7_-_66915756 | 1.98 |
ENSMUST00000207715.2
|
Mef2a
|
myocyte enhancer factor 2A |
| chr15_+_59246080 | 1.88 |
ENSMUST00000168722.3
|
Nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr6_+_54241830 | 1.86 |
ENSMUST00000146114.8
|
Chn2
|
chimerin 2 |
| chr4_+_51216645 | 1.84 |
ENSMUST00000166749.2
ENSMUST00000156384.4 |
Cylc2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr1_+_167445815 | 1.83 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr19_-_11142662 | 1.75 |
ENSMUST00000181567.8
ENSMUST00000180678.8 ENSMUST00000078770.5 |
Ms4a19
|
membrane-spanning 4-domains, subfamily A, member 19 |
| chr12_-_46767619 | 1.73 |
ENSMUST00000219886.2
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr5_+_35146727 | 1.72 |
ENSMUST00000114284.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr15_-_54783357 | 1.72 |
ENSMUST00000167541.3
ENSMUST00000171545.9 ENSMUST00000041591.16 ENSMUST00000173516.8 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr10_+_5589210 | 1.71 |
ENSMUST00000019906.6
|
Vip
|
vasoactive intestinal polypeptide |
| chr2_-_37537224 | 1.67 |
ENSMUST00000028279.10
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr16_-_10606513 | 1.66 |
ENSMUST00000051297.9
|
Tnp2
|
transition protein 2 |
| chr7_+_44926925 | 1.63 |
ENSMUST00000210861.2
|
Slc6a21
|
solute carrier family 6 member 21 |
| chr18_+_37630044 | 1.62 |
ENSMUST00000059571.7
|
Pcdhb19
|
protocadherin beta 19 |
| chr6_-_90758954 | 1.62 |
ENSMUST00000238821.2
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr5_-_29583300 | 1.62 |
ENSMUST00000196321.5
ENSMUST00000200564.5 ENSMUST00000055195.11 ENSMUST00000198105.5 ENSMUST00000179191.6 |
Lmbr1
|
limb region 1 |
| chr11_+_71640739 | 1.60 |
ENSMUST00000150531.2
|
Wscd1
|
WSC domain containing 1 |
| chr9_-_46231282 | 1.60 |
ENSMUST00000159565.8
|
4931429L15Rik
|
RIKEN cDNA 4931429L15 gene |
| chr13_+_83652280 | 1.60 |
ENSMUST00000199450.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr18_-_10706701 | 1.56 |
ENSMUST00000002549.9
ENSMUST00000117726.9 ENSMUST00000117828.9 |
Abhd3
|
abhydrolase domain containing 3 |
| chr4_+_43406435 | 1.55 |
ENSMUST00000098106.9
ENSMUST00000139198.2 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr9_-_50650663 | 1.51 |
ENSMUST00000117093.2
ENSMUST00000121634.8 |
Dixdc1
|
DIX domain containing 1 |
| chr2_-_42543069 | 1.48 |
ENSMUST00000203080.3
|
Lrp1b
|
low density lipoprotein-related protein 1B |
| chr17_+_13279986 | 1.43 |
ENSMUST00000086787.11
ENSMUST00000116666.8 ENSMUST00000232990.2 ENSMUST00000233511.2 |
Tcp10b
|
t-complex protein 10b |
| chr13_+_83723255 | 1.41 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr1_-_172722589 | 1.39 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr13_-_90237179 | 1.39 |
ENSMUST00000161396.2
|
Xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr2_-_164041997 | 1.38 |
ENSMUST00000063251.3
|
Wfdc15a
|
WAP four-disulfide core domain 15A |
| chr18_+_37544717 | 1.38 |
ENSMUST00000051126.4
|
Pcdhb10
|
protocadherin beta 10 |
| chr4_-_119241002 | 1.37 |
ENSMUST00000238721.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr14_-_4506874 | 1.34 |
ENSMUST00000224934.2
|
Thrb
|
thyroid hormone receptor beta |
| chr3_+_137329433 | 1.28 |
ENSMUST00000053855.8
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr7_-_104991477 | 1.27 |
ENSMUST00000213290.2
|
Olfr691
|
olfactory receptor 691 |
| chr8_+_46081213 | 1.26 |
ENSMUST00000130850.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr2_-_23939401 | 1.26 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr3_-_92050043 | 1.26 |
ENSMUST00000197811.2
ENSMUST00000029535.6 |
4930511M18Rik
Lelp1
|
RIKEN cDNA 4930511M18 gene late cornified envelope-like proline-rich 1 |
| chr18_+_52779281 | 1.25 |
ENSMUST00000118724.8
ENSMUST00000091904.6 |
1700034E13Rik
|
RIKEN cDNA 1700034E13 gene |
| chr5_+_35146880 | 1.24 |
ENSMUST00000114285.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chrX_+_77413242 | 1.24 |
ENSMUST00000035626.8
|
4930480E11Rik
|
RIKEN cDNA 4930480E11 gene |
| chr9_+_30338329 | 1.23 |
ENSMUST00000164099.3
|
Snx19
|
sorting nexin 19 |
| chr1_+_179788675 | 1.22 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr6_-_131655849 | 1.19 |
ENSMUST00000076756.3
|
Tas2r106
|
taste receptor, type 2, member 106 |
| chr3_-_85909798 | 1.19 |
ENSMUST00000061343.4
|
Prss48
|
protease, serine 48 |
| chrX_+_111513971 | 1.15 |
ENSMUST00000071814.13
|
Zfp711
|
zinc finger protein 711 |
| chr19_-_45580258 | 1.14 |
ENSMUST00000160003.8
ENSMUST00000162879.8 |
Fbxw4
|
F-box and WD-40 domain protein 4 |
| chr13_+_23433408 | 1.12 |
ENSMUST00000091719.2
|
Vmn1r223
|
vomeronasal 1 receptor 223 |
| chr19_-_39637489 | 1.11 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr10_+_40446326 | 1.10 |
ENSMUST00000078314.14
|
Slc22a16
|
solute carrier family 22 (organic cation transporter), member 16 |
| chr18_-_3281752 | 1.08 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr4_-_14796052 | 1.07 |
ENSMUST00000108276.2
ENSMUST00000023917.8 |
Lrrc69
|
leucine rich repeat containing 69 |
| chr3_+_66892979 | 1.01 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr12_-_84031622 | 0.99 |
ENSMUST00000164935.3
|
Heatr4
|
HEAT repeat containing 4 |
| chr19_+_6111204 | 0.99 |
ENSMUST00000162726.5
|
Znhit2
|
zinc finger, HIT domain containing 2 |
| chr11_+_85202990 | 0.99 |
ENSMUST00000127717.2
|
Ppm1d
|
protein phosphatase 1D magnesium-dependent, delta isoform |
| chr12_-_76293459 | 0.98 |
ENSMUST00000219327.2
ENSMUST00000021453.6 ENSMUST00000218426.2 |
Tex21
|
testis expressed gene 21 |
| chr1_-_127782735 | 0.98 |
ENSMUST00000208183.3
|
Map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr18_+_37646674 | 0.97 |
ENSMUST00000061405.6
|
Pcdhb21
|
protocadherin beta 21 |
| chr10_-_112764879 | 0.97 |
ENSMUST00000099276.4
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr6_-_56878854 | 0.96 |
ENSMUST00000101367.9
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr16_+_88954838 | 0.96 |
ENSMUST00000062524.6
|
Gm9789
|
predicted gene 9789 |
| chrX_+_106836189 | 0.95 |
ENSMUST00000101292.9
|
Tent5d
|
terminal nucleotidyltransferase 5D |
| chr8_+_47439916 | 0.95 |
ENSMUST00000039840.15
|
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr7_-_130748035 | 0.94 |
ENSMUST00000070980.4
|
4933402N03Rik
|
RIKEN cDNA 4933402N03 gene |
| chr6_+_36364990 | 0.94 |
ENSMUST00000172278.8
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr8_+_47439948 | 0.90 |
ENSMUST00000119686.2
|
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr13_-_48746836 | 0.90 |
ENSMUST00000238995.2
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr13_+_18901459 | 0.88 |
ENSMUST00000072961.6
|
Vps41
|
VPS41 HOPS complex subunit |
| chr13_+_110063364 | 0.87 |
ENSMUST00000117420.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr6_-_39397334 | 0.86 |
ENSMUST00000031985.13
|
Mkrn1
|
makorin, ring finger protein, 1 |
| chr1_+_58249556 | 0.86 |
ENSMUST00000040442.6
|
Aox4
|
aldehyde oxidase 4 |
| chr2_+_69621312 | 0.85 |
ENSMUST00000180290.2
|
Phospho2
|
phosphatase, orphan 2 |
| chr8_+_46945826 | 0.84 |
ENSMUST00000110371.8
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr1_-_162913210 | 0.84 |
ENSMUST00000096608.5
|
Mroh9
|
maestro heat-like repeat family member 9 |
| chr18_+_37651393 | 0.83 |
ENSMUST00000097609.3
|
Pcdhb22
|
protocadherin beta 22 |
| chr19_+_39499288 | 0.82 |
ENSMUST00000025968.5
|
Cyp2c39
|
cytochrome P450, family 2, subfamily c, polypeptide 39 |
| chrX_+_110154017 | 0.82 |
ENSMUST00000210720.3
|
Cylc1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr11_+_58592529 | 0.82 |
ENSMUST00000076965.2
|
Olfr319
|
olfactory receptor 319 |
| chr8_-_27713841 | 0.82 |
ENSMUST00000209299.2
|
Got1l1
|
glutamic-oxaloacetic transaminase 1-like 1 |
| chr1_-_80642969 | 0.82 |
ENSMUST00000190983.7
ENSMUST00000191449.2 |
Dock10
|
dedicator of cytokinesis 10 |
| chr11_-_12362136 | 0.82 |
ENSMUST00000174874.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr2_+_164664920 | 0.81 |
ENSMUST00000132282.2
|
Zswim1
|
zinc finger SWIM-type containing 1 |
| chr1_+_179788037 | 0.80 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_+_87362140 | 0.78 |
ENSMUST00000217113.2
|
Olfr153
|
olfactory receptor 153 |
| chr2_-_114031897 | 0.76 |
ENSMUST00000050668.4
|
Zfp770
|
zinc finger protein 770 |
| chr14_-_50521663 | 0.73 |
ENSMUST00000213701.2
|
Olfr732
|
olfactory receptor 732 |
| chr15_+_37425798 | 0.73 |
ENSMUST00000022897.2
|
4930447A16Rik
|
RIKEN cDNA 4930447A16 gene |
| chr3_-_63391300 | 0.73 |
ENSMUST00000192926.2
|
Strit1
|
small transmembrane regulator of ion transport 1 |
| chr11_+_49327451 | 0.71 |
ENSMUST00000215226.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr11_-_100653754 | 0.71 |
ENSMUST00000107360.3
ENSMUST00000055083.4 |
Hcrt
|
hypocretin |
| chr7_-_14180576 | 0.71 |
ENSMUST00000125941.3
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr17_+_38485977 | 0.70 |
ENSMUST00000074883.2
|
Olfr134
|
olfactory receptor 134 |
| chr5_-_6926523 | 0.69 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr12_+_71936500 | 0.69 |
ENSMUST00000221317.2
|
Daam1
|
dishevelled associated activator of morphogenesis 1 |
| chr1_+_136059101 | 0.69 |
ENSMUST00000075164.11
|
Kif21b
|
kinesin family member 21B |
| chr11_+_51895166 | 0.64 |
ENSMUST00000109076.2
|
Cdkl3
|
cyclin-dependent kinase-like 3 |
| chr9_+_38179147 | 0.64 |
ENSMUST00000212156.3
|
Olfr895
|
olfactory receptor 895 |
| chr18_+_87774402 | 0.62 |
ENSMUST00000091776.7
|
Gm5096
|
predicted gene 5096 |
| chr2_-_77349909 | 0.61 |
ENSMUST00000111830.9
|
Zfp385b
|
zinc finger protein 385B |
| chr1_+_74401267 | 0.61 |
ENSMUST00000097697.8
|
Catip
|
ciliogenesis associated TTC17 interacting protein |
| chr2_-_34716083 | 0.60 |
ENSMUST00000113077.8
ENSMUST00000028220.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr12_-_16639721 | 0.59 |
ENSMUST00000221146.2
|
Lpin1
|
lipin 1 |
| chr11_+_3280401 | 0.59 |
ENSMUST00000045153.11
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr15_-_76906150 | 0.57 |
ENSMUST00000230031.2
|
Mb
|
myoglobin |
| chr2_+_87610895 | 0.56 |
ENSMUST00000215394.2
|
Olfr152
|
olfactory receptor 152 |
| chr3_+_94305824 | 0.55 |
ENSMUST00000050975.6
|
Lingo4
|
leucine rich repeat and Ig domain containing 4 |
| chr9_+_51027300 | 0.55 |
ENSMUST00000114431.4
ENSMUST00000213117.2 |
Btg4
|
BTG anti-proliferation factor 4 |
| chr16_-_90866032 | 0.54 |
ENSMUST00000035689.8
ENSMUST00000114076.2 |
4932438H23Rik
|
RIKEN cDNA 4932438H23 gene |
| chr10_-_117628565 | 0.52 |
ENSMUST00000167943.8
ENSMUST00000064848.7 |
Nup107
|
nucleoporin 107 |
| chr16_+_38182569 | 0.52 |
ENSMUST00000023494.13
ENSMUST00000114739.2 |
Popdc2
|
popeye domain containing 2 |
| chr14_-_50538979 | 0.51 |
ENSMUST00000216195.2
ENSMUST00000214372.2 ENSMUST00000214756.2 |
Olfr733
|
olfactory receptor 733 |
| chr14_-_57353340 | 0.51 |
ENSMUST00000159455.2
|
Gm4491
|
predicted gene 4491 |
| chr1_-_132157505 | 0.51 |
ENSMUST00000086544.5
|
Gm10188
|
predicted gene 10188 |
| chr2_-_89172742 | 0.51 |
ENSMUST00000216561.2
|
Olfr1233
|
olfactory receptor 1233 |
| chr17_+_6130061 | 0.50 |
ENSMUST00000039487.10
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr2_-_34716199 | 0.50 |
ENSMUST00000113075.8
ENSMUST00000113080.9 ENSMUST00000091020.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr10_-_30647836 | 0.50 |
ENSMUST00000215926.2
ENSMUST00000213836.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr5_-_123804745 | 0.48 |
ENSMUST00000149410.2
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr4_+_33031342 | 0.48 |
ENSMUST00000124992.8
|
Ube2j1
|
ubiquitin-conjugating enzyme E2J 1 |
| chr9_-_40915858 | 0.47 |
ENSMUST00000188848.8
ENSMUST00000034519.13 |
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr14_+_21800593 | 0.43 |
ENSMUST00000022292.10
|
Samd8
|
sterile alpha motif domain containing 8 |
| chr5_+_117378510 | 0.41 |
ENSMUST00000111975.3
|
Taok3
|
TAO kinase 3 |
| chr6_-_23655130 | 0.41 |
ENSMUST00000104979.2
|
Rnf148
|
ring finger protein 148 |
| chr5_+_146016064 | 0.39 |
ENSMUST00000035571.10
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chrM_+_3906 | 0.37 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chrX_+_10118600 | 0.36 |
ENSMUST00000115528.3
|
Otc
|
ornithine transcarbamylase |
| chr7_-_73187369 | 0.34 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr8_-_43981143 | 0.33 |
ENSMUST00000080135.5
|
Adam26b
|
a disintegrin and metallopeptidase domain 26B |
| chr10_-_129023288 | 0.33 |
ENSMUST00000072976.4
|
Olfr773
|
olfactory receptor 773 |
| chr5_-_108943211 | 0.32 |
ENSMUST00000004943.2
|
Tmed11
|
transmembrane p24 trafficking protein 11 |
| chr9_-_40915895 | 0.32 |
ENSMUST00000180384.3
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr7_-_103721076 | 0.31 |
ENSMUST00000213184.2
ENSMUST00000213991.2 |
Olfr644
|
olfactory receptor 644 |
| chr11_-_101308441 | 0.29 |
ENSMUST00000070395.9
|
Aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr1_-_160079007 | 0.29 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr1_-_156767123 | 0.29 |
ENSMUST00000189316.7
ENSMUST00000190648.7 ENSMUST00000172057.8 ENSMUST00000191605.7 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr12_+_25024913 | 0.28 |
ENSMUST00000066652.7
ENSMUST00000220459.2 ENSMUST00000222941.2 |
Kidins220
|
kinase D-interacting substrate 220 |
| chr4_+_33132502 | 0.28 |
ENSMUST00000029947.6
|
Gabrr1
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 1 |
| chr7_-_10292412 | 0.27 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68 |
| chr7_-_102774821 | 0.27 |
ENSMUST00000211075.3
ENSMUST00000215304.2 ENSMUST00000213281.2 |
Olfr586
|
olfactory receptor 586 |
| chr13_-_56696222 | 0.27 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr10_-_13053733 | 0.27 |
ENSMUST00000019954.6
|
Zc2hc1b
|
zinc finger, C2HC-type containing 1B |
| chr9_+_37766116 | 0.26 |
ENSMUST00000086063.4
|
Olfr877
|
olfactory receptor 877 |
| chr16_+_44215136 | 0.26 |
ENSMUST00000099742.9
|
Cfap44
|
cilia and flagella associated protein 44 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) |
| 1.1 | 4.3 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 1.0 | 6.9 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.9 | 3.7 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.9 | 7.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.8 | 4.6 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.7 | 5.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.7 | 4.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.6 | 7.6 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.6 | 3.0 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.6 | 4.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.6 | 2.8 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.5 | 1.4 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.5 | 2.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.4 | 2.4 | GO:0036482 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of macromitophagy(GO:1901526) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.4 | 8.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 2.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.4 | 2.9 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.3 | 2.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 1.9 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.3 | 6.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 0.8 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.3 | 3.9 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.3 | 1.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 1.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.2 | 1.7 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 1.7 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 1.8 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 0.8 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.2 | 4.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 0.6 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.2 | 9.4 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 0.9 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.2 | 0.9 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.2 | 4.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.2 | 1.7 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.1 | 2.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 4.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.7 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.8 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 1.5 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 2.9 | GO:1903831 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.5 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.5 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 1.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.5 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 1.4 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 3.0 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 3.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 1.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.8 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 2.0 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 4.9 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 2.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.7 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 1.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.5 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.6 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 1.8 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 1.6 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 2.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 1.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 2.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 5.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 2.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 1.0 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.2 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 1.1 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 2.3 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.6 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 1.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.9 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.6 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.7 | 6.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.5 | 4.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 4.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 8.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 3.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 3.3 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 6.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.2 | 2.8 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.2 | 12.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 0.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 2.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 1.9 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 3.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.9 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.8 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 2.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 5.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 4.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.5 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 4.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 8.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 3.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 7.6 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 2.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 2.4 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 1.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 12.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.0 | 9.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.9 | 3.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.9 | 2.8 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.8 | 4.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.8 | 8.7 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.6 | 4.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 3.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.4 | 4.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 1.7 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.3 | 4.0 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.3 | 2.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 2.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 7.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 2.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 1.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.3 | 7.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 7.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 0.9 | GO:1990763 | G-protein coupled acetylcholine receptor activity(GO:0016907) arrestin family protein binding(GO:1990763) |
| 0.2 | 3.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 1.4 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 1.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.2 | 0.8 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.2 | 0.8 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 1.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 2.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 1.8 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 1.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 1.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 0.9 | GO:0030151 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) |
| 0.2 | 0.8 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.1 | 2.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.6 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 1.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 1.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.4 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 2.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.2 | GO:0001132 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.1 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.9 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 3.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 3.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 2.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 5.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 2.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 1.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.6 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 3.3 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 4.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 6.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 3.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 6.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 4.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 13.7 | REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | Genes involved in MAPK targets/ Nuclear events mediated by MAP kinases |
| 0.1 | 2.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 6.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 4.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 2.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 1.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 2.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 2.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 3.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 4.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |