Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
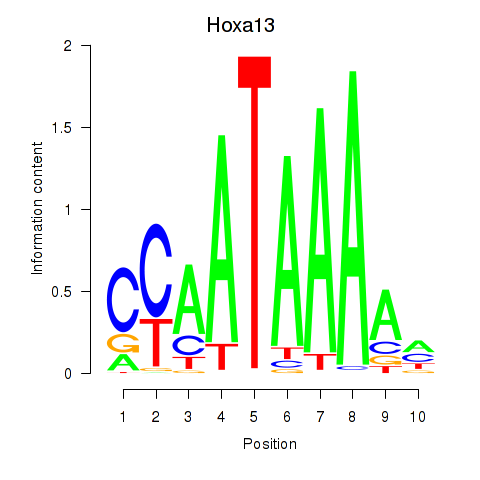
Results for Hoxa13
Z-value: 1.35
Transcription factors associated with Hoxa13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa13
|
ENSMUSG00000038203.22 | Hoxa13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa13 | mm39_v1_chr6_-_52237765_52237788 | -0.04 | 7.2e-01 | Click! |
Activity profile of Hoxa13 motif
Sorted Z-values of Hoxa13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_110305672 | 13.25 |
ENSMUST00000074898.8
|
Hp
|
haptoglobin |
| chr4_-_61592331 | 11.57 |
ENSMUST00000098040.4
|
Mup18
|
major urinary protein 18 |
| chr19_+_40078132 | 11.55 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr4_-_61259801 | 11.45 |
ENSMUST00000125461.8
|
Mup14
|
major urinary protein 14 |
| chr4_-_61259997 | 11.14 |
ENSMUST00000071005.9
ENSMUST00000075206.12 |
Mup14
|
major urinary protein 14 |
| chr3_-_10273628 | 9.67 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr4_-_60070411 | 8.72 |
ENSMUST00000079697.10
ENSMUST00000125282.2 ENSMUST00000166098.8 |
Mup7
|
major urinary protein 7 |
| chr4_-_61184270 | 8.69 |
ENSMUST00000072678.6
ENSMUST00000098042.10 |
Mup13
|
major urinary protein 13 |
| chr4_-_61357980 | 8.51 |
ENSMUST00000095049.5
|
Mup15
|
major urinary protein 15 |
| chr4_-_60538151 | 8.31 |
ENSMUST00000098047.3
|
Mup10
|
major urinary protein 10 |
| chr4_-_60377932 | 8.10 |
ENSMUST00000107506.9
ENSMUST00000122381.8 ENSMUST00000118759.8 ENSMUST00000132829.3 |
Mup9
|
major urinary protein 9 |
| chr4_-_60139857 | 8.01 |
ENSMUST00000107490.5
ENSMUST00000074700.9 |
Mup2
|
major urinary protein 2 |
| chr4_+_98919183 | 8.00 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr2_-_69172944 | 7.96 |
ENSMUST00000102709.8
ENSMUST00000102710.10 ENSMUST00000180142.2 |
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr4_-_60618357 | 7.93 |
ENSMUST00000084544.5
ENSMUST00000098046.10 |
Mup11
|
major urinary protein 11 |
| chr4_-_61437704 | 7.90 |
ENSMUST00000095051.6
ENSMUST00000107483.8 |
Mup16
|
major urinary protein 16 |
| chr5_-_89583469 | 7.75 |
ENSMUST00000200534.2
|
Gc
|
vitamin D binding protein |
| chr15_+_6474808 | 7.73 |
ENSMUST00000022749.17
ENSMUST00000239466.2 |
C9
|
complement component 9 |
| chr4_-_61514108 | 7.69 |
ENSMUST00000107484.2
|
Mup17
|
major urinary protein 17 |
| chr4_-_60222580 | 7.63 |
ENSMUST00000095058.5
ENSMUST00000163931.8 |
Mup8
|
major urinary protein 8 |
| chr4_-_60777462 | 7.44 |
ENSMUST00000211875.2
|
Mup22
|
major urinary protein 22 |
| chr7_+_25872836 | 7.31 |
ENSMUST00000082214.5
|
Cyp2b9
|
cytochrome P450, family 2, subfamily b, polypeptide 9 |
| chr10_+_97315465 | 6.84 |
ENSMUST00000105287.11
|
Dcn
|
decorin |
| chr4_-_60697274 | 6.68 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr19_+_12610870 | 6.49 |
ENSMUST00000119960.2
|
Glyat
|
glycine-N-acyltransferase |
| chr3_+_20039775 | 6.29 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin |
| chr4_-_60457902 | 6.28 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr16_+_22769844 | 6.10 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr7_-_30643444 | 5.98 |
ENSMUST00000062620.9
|
Hamp
|
hepcidin antimicrobial peptide |
| chr7_+_119206233 | 5.98 |
ENSMUST00000126367.8
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr19_+_12610668 | 5.92 |
ENSMUST00000044976.12
|
Glyat
|
glycine-N-acyltransferase |
| chr13_-_32967937 | 5.77 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chrY_-_1286623 | 5.61 |
ENSMUST00000091190.12
|
Ddx3y
|
DEAD box helicase 3, Y-linked |
| chr7_-_30623592 | 5.59 |
ENSMUST00000217812.2
ENSMUST00000074671.9 |
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr8_-_25066313 | 5.50 |
ENSMUST00000121992.2
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr8_+_15107646 | 5.39 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
| chr13_+_93810911 | 5.31 |
ENSMUST00000048001.8
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr9_-_105022272 | 5.28 |
ENSMUST00000190661.2
ENSMUST00000035180.5 |
Nudt16l2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 2 |
| chr8_-_106670014 | 5.27 |
ENSMUST00000038896.8
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr16_+_22769822 | 5.22 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr10_-_28862289 | 5.11 |
ENSMUST00000152363.8
ENSMUST00000015663.7 |
2310057J18Rik
|
RIKEN cDNA 2310057J18 gene |
| chr15_+_101371353 | 5.07 |
ENSMUST00000088049.5
|
Krt86
|
keratin 86 |
| chr11_+_96085118 | 5.07 |
ENSMUST00000062709.4
|
Hoxb13
|
homeobox B13 |
| chr10_+_4561974 | 4.83 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr4_-_108075119 | 4.81 |
ENSMUST00000223127.2
ENSMUST00000043793.7 ENSMUST00000106690.9 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr14_-_47426863 | 4.80 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr13_+_81031512 | 4.68 |
ENSMUST00000099356.10
|
Arrdc3
|
arrestin domain containing 3 |
| chr3_+_98187743 | 4.64 |
ENSMUST00000120541.8
ENSMUST00000090746.3 |
Hmgcs2
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 |
| chr1_-_71692320 | 4.62 |
ENSMUST00000186940.7
ENSMUST00000188894.7 ENSMUST00000188674.7 ENSMUST00000189821.7 ENSMUST00000187938.7 ENSMUST00000190780.7 ENSMUST00000186736.2 ENSMUST00000055226.13 ENSMUST00000186129.7 |
Fn1
|
fibronectin 1 |
| chr2_-_91025441 | 4.56 |
ENSMUST00000002177.9
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr5_-_69699932 | 4.54 |
ENSMUST00000202423.2
|
Yipf7
|
Yip1 domain family, member 7 |
| chr7_+_25760922 | 4.50 |
ENSMUST00000005669.9
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr5_-_69699965 | 4.46 |
ENSMUST00000031045.10
|
Yipf7
|
Yip1 domain family, member 7 |
| chr3_-_146321341 | 4.44 |
ENSMUST00000200633.2
|
Dnase2b
|
deoxyribonuclease II beta |
| chr7_+_127399776 | 4.42 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr2_-_91025492 | 4.28 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr7_+_101750943 | 4.26 |
ENSMUST00000033300.4
|
Art1
|
ADP-ribosyltransferase 1 |
| chrX_+_59044796 | 4.23 |
ENSMUST00000033477.5
|
F9
|
coagulation factor IX |
| chr8_-_5155347 | 4.21 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr10_+_111896087 | 4.20 |
ENSMUST00000073617.5
|
Glipr1l1
|
GLI pathogenesis-related 1 like 1 |
| chr3_-_88317601 | 4.15 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr12_-_55033113 | 4.15 |
ENSMUST00000038926.13
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr12_-_72675624 | 4.13 |
ENSMUST00000208039.2
ENSMUST00000207585.2 |
Gm4756
|
predicted gene 4756 |
| chr10_+_52267702 | 4.11 |
ENSMUST00000067085.7
|
Nepn
|
nephrocan |
| chr1_+_175459559 | 4.07 |
ENSMUST00000040250.15
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr5_-_145946408 | 4.07 |
ENSMUST00000138870.2
ENSMUST00000068317.13 |
Cyp3a25
|
cytochrome P450, family 3, subfamily a, polypeptide 25 |
| chr12_+_113038376 | 4.06 |
ENSMUST00000109729.3
|
Tex22
|
testis expressed gene 22 |
| chr7_-_19411866 | 4.04 |
ENSMUST00000142352.9
|
Apoc2
|
apolipoprotein C-II |
| chr12_-_104120105 | 4.01 |
ENSMUST00000085050.4
|
Serpina3c
|
serine (or cysteine) peptidase inhibitor, clade A, member 3C |
| chr5_+_146016064 | 4.01 |
ENSMUST00000035571.10
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr8_-_94006345 | 4.00 |
ENSMUST00000034178.9
|
Ces1f
|
carboxylesterase 1F |
| chr2_+_74528071 | 3.99 |
ENSMUST00000059272.10
|
Hoxd9
|
homeobox D9 |
| chr2_-_91025380 | 3.84 |
ENSMUST00000111356.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr1_+_175459735 | 3.79 |
ENSMUST00000097458.4
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr14_+_26616514 | 3.77 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr2_-_164080358 | 3.75 |
ENSMUST00000044953.3
|
Svs2
|
seminal vesicle secretory protein 2 |
| chr2_-_164080172 | 3.74 |
ENSMUST00000109374.2
|
Svs2
|
seminal vesicle secretory protein 2 |
| chr1_-_192946359 | 3.71 |
ENSMUST00000161737.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr2_-_32341408 | 3.70 |
ENSMUST00000028160.15
ENSMUST00000113310.9 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr12_+_34007645 | 3.70 |
ENSMUST00000049089.7
|
Twist1
|
twist basic helix-loop-helix transcription factor 1 |
| chr2_+_74535242 | 3.66 |
ENSMUST00000019749.4
|
Hoxd8
|
homeobox D8 |
| chr15_-_101422054 | 3.66 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chr5_-_87485023 | 3.64 |
ENSMUST00000031195.3
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr5_+_90708962 | 3.63 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr12_-_72132168 | 3.63 |
ENSMUST00000019862.3
|
L3hypdh
|
L-3-hydroxyproline dehydratase (trans-) |
| chr11_+_96214078 | 3.56 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr4_-_3938352 | 3.55 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr13_-_24098981 | 3.51 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr3_+_19562771 | 3.43 |
ENSMUST00000029132.11
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr16_+_34470290 | 3.38 |
ENSMUST00000148562.8
|
Ropn1
|
ropporin, rhophilin associated protein 1 |
| chr2_+_74534959 | 3.25 |
ENSMUST00000151380.2
|
Hoxd8
|
homeobox D8 |
| chr13_-_24098951 | 3.23 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr8_-_25528972 | 3.22 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4 |
| chr1_-_97055931 | 3.21 |
ENSMUST00000027569.14
|
Slco6c1
|
solute carrier organic anion transporter family, member 6c1 |
| chr19_-_8109346 | 3.21 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr2_-_91025208 | 3.21 |
ENSMUST00000111355.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr19_-_6117815 | 3.20 |
ENSMUST00000162575.8
ENSMUST00000159084.8 ENSMUST00000161718.8 ENSMUST00000162810.8 ENSMUST00000025713.12 ENSMUST00000113543.9 ENSMUST00000160417.8 ENSMUST00000161528.2 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr12_+_113038098 | 3.19 |
ENSMUST00000012355.14
ENSMUST00000146107.8 |
Tex22
|
testis expressed gene 22 |
| chr12_-_55033130 | 3.19 |
ENSMUST00000173433.8
ENSMUST00000173803.2 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr2_+_74535489 | 3.18 |
ENSMUST00000074721.6
|
Hoxd8
|
homeobox D8 |
| chr11_+_70548022 | 3.17 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr9_-_106125055 | 3.14 |
ENSMUST00000074082.13
|
Alas1
|
aminolevulinic acid synthase 1 |
| chr3_-_122828592 | 3.14 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr3_+_96736774 | 3.13 |
ENSMUST00000138014.8
|
Pdzk1
|
PDZ domain containing 1 |
| chr7_-_136916123 | 3.09 |
ENSMUST00000106118.10
ENSMUST00000168203.2 ENSMUST00000169486.9 ENSMUST00000033378.13 |
Ebf3
|
early B cell factor 3 |
| chr1_-_97056017 | 3.08 |
ENSMUST00000189547.2
|
Slco6c1
|
solute carrier organic anion transporter family, member 6c1 |
| chr6_-_52217821 | 3.06 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr3_+_96127174 | 3.06 |
ENSMUST00000073115.5
|
H2ac21
|
H2A clustered histone 21 |
| chr18_-_3281752 | 3.04 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr4_-_120966396 | 3.00 |
ENSMUST00000106268.4
|
Tmco2
|
transmembrane and coiled-coil domains 2 |
| chr7_+_134870237 | 2.99 |
ENSMUST00000210697.2
ENSMUST00000097983.5 |
Nps
|
neuropeptide S |
| chr9_-_106124917 | 2.98 |
ENSMUST00000112524.9
ENSMUST00000219129.2 |
Alas1
|
aminolevulinic acid synthase 1 |
| chr10_+_116137277 | 2.95 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr7_+_80707328 | 2.91 |
ENSMUST00000107348.2
|
Alpk3
|
alpha-kinase 3 |
| chr3_-_104684798 | 2.91 |
ENSMUST00000139783.2
|
Tafa3
|
TAFA chemokine like family member 3 |
| chr3_+_19562753 | 2.89 |
ENSMUST00000118968.8
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr16_+_26281885 | 2.87 |
ENSMUST00000161053.8
ENSMUST00000115302.2 |
Cldn16
|
claudin 16 |
| chr11_+_96920751 | 2.86 |
ENSMUST00000021249.11
|
Scrn2
|
secernin 2 |
| chr3_+_96736600 | 2.86 |
ENSMUST00000135031.8
|
Pdzk1
|
PDZ domain containing 1 |
| chr1_-_72251466 | 2.84 |
ENSMUST00000048860.9
|
Mreg
|
melanoregulin |
| chr12_-_57244096 | 2.82 |
ENSMUST00000044634.12
|
Slc25a21
|
solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 |
| chr13_-_64460491 | 2.80 |
ENSMUST00000222570.2
ENSMUST00000220895.2 |
Prxl2c
|
peroxiredoxin like 2C |
| chr4_-_34756222 | 2.78 |
ENSMUST00000095129.3
|
Gm136
|
predicted gene 136 |
| chr3_+_20011201 | 2.73 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr5_-_145406533 | 2.72 |
ENSMUST00000031633.5
|
Cyp3a16
|
cytochrome P450, family 3, subfamily a, polypeptide 16 |
| chr15_-_101441254 | 2.68 |
ENSMUST00000023720.8
|
Krt84
|
keratin 84 |
| chr10_+_17672004 | 2.67 |
ENSMUST00000037964.7
|
Txlnb
|
taxilin beta |
| chr9_+_51958453 | 2.66 |
ENSMUST00000163153.9
|
Rdx
|
radixin |
| chrX_-_74205226 | 2.64 |
ENSMUST00000165080.2
|
Smim9
|
small integral membrane protein 9 |
| chr11_-_88608958 | 2.63 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr3_+_20011405 | 2.63 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr2_-_35257741 | 2.63 |
ENSMUST00000028243.2
|
4930568D16Rik
|
RIKEN cDNA 4930568D16 gene |
| chr7_-_98829474 | 2.60 |
ENSMUST00000207611.2
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr7_-_103799746 | 2.59 |
ENSMUST00000059121.5
|
Ubqlnl
|
ubiquilin-like |
| chr6_+_52290415 | 2.57 |
ENSMUST00000031787.8
ENSMUST00000129243.3 |
Evx1
|
even-skipped homeobox 1 |
| chr12_-_86773160 | 2.55 |
ENSMUST00000021682.9
|
Angel1
|
angel homolog 1 |
| chr12_-_72675670 | 2.53 |
ENSMUST00000209038.2
|
Gm4756
|
predicted gene 4756 |
| chr18_+_39906550 | 2.49 |
ENSMUST00000063219.3
|
Pabpc2
|
poly(A) binding protein, cytoplasmic 2 |
| chr6_+_82018604 | 2.46 |
ENSMUST00000042974.15
|
Eva1a
|
eva-1 homolog A (C. elegans) |
| chr6_-_113577606 | 2.45 |
ENSMUST00000035870.5
|
Fancd2os
|
Fancd2 opposite strand |
| chr16_+_34471558 | 2.38 |
ENSMUST00000023530.5
|
Ropn1
|
ropporin, rhophilin associated protein 1 |
| chr3_+_19241484 | 2.38 |
ENSMUST00000130806.8
ENSMUST00000117529.8 ENSMUST00000119865.8 |
Mtfr1
|
mitochondrial fission regulator 1 |
| chr8_+_57774010 | 2.38 |
ENSMUST00000040104.5
|
Hand2
|
heart and neural crest derivatives expressed 2 |
| chr14_-_52150804 | 2.38 |
ENSMUST00000004673.15
ENSMUST00000111632.5 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr8_-_25215856 | 2.37 |
ENSMUST00000033958.15
|
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr1_+_181404124 | 2.36 |
ENSMUST00000208001.2
|
Dnah14
|
dynein, axonemal, heavy chain 14 |
| chrM_+_3906 | 2.35 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr14_+_41765952 | 2.35 |
ENSMUST00000168972.2
ENSMUST00000064162.11 |
1700024B05Rik
|
RIKEN cDNA 1700024B05 gene |
| chr1_-_173594475 | 2.35 |
ENSMUST00000111214.4
|
Ifi204
|
interferon activated gene 204 |
| chr7_-_19432129 | 2.34 |
ENSMUST00000172808.2
ENSMUST00000174191.2 |
Apoe
|
apolipoprotein E |
| chr11_-_99996452 | 2.33 |
ENSMUST00000107416.3
|
Krt36
|
keratin 36 |
| chr1_-_121260274 | 2.33 |
ENSMUST00000161068.2
|
Insig2
|
insulin induced gene 2 |
| chr12_+_113038397 | 2.30 |
ENSMUST00000155492.2
|
Tex22
|
testis expressed gene 22 |
| chr2_+_85868891 | 2.26 |
ENSMUST00000218397.2
|
Olfr1033
|
olfactory receptor 1033 |
| chr5_-_87739442 | 2.25 |
ENSMUST00000031201.9
|
Sult1e1
|
sulfotransferase family 1E, member 1 |
| chr12_-_103623354 | 2.24 |
ENSMUST00000152517.2
|
Serpina6
|
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
| chr10_-_53252210 | 2.24 |
ENSMUST00000095691.7
|
Cep85l
|
centrosomal protein 85-like |
| chr14_+_33775423 | 2.24 |
ENSMUST00000058725.5
|
Antxrl
|
anthrax toxin receptor-like |
| chr3_+_20011251 | 2.23 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chrX_+_105964224 | 2.21 |
ENSMUST00000060576.8
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr2_-_180284468 | 2.19 |
ENSMUST00000037877.11
|
Tcfl5
|
transcription factor-like 5 (basic helix-loop-helix) |
| chr6_+_145561483 | 2.16 |
ENSMUST00000087445.7
|
Tuba3b
|
tubulin, alpha 3B |
| chr15_-_101459092 | 2.16 |
ENSMUST00000023713.10
|
Krt82
|
keratin 82 |
| chr8_+_27937128 | 2.13 |
ENSMUST00000209669.2
|
Gm45861
|
predicted gene 45861 |
| chr8_-_79975199 | 2.12 |
ENSMUST00000034109.6
|
1700011L22Rik
|
RIKEN cDNA 1700011L22 gene |
| chr10_-_129738595 | 2.11 |
ENSMUST00000071557.2
|
Olfr815
|
olfactory receptor 815 |
| chr8_-_96302083 | 2.07 |
ENSMUST00000213096.2
ENSMUST00000052690.13 |
Prss54
|
protease, serine 54 |
| chr13_+_34918820 | 2.06 |
ENSMUST00000039605.8
|
Fam50b
|
family with sequence similarity 50, member B |
| chrX_-_133442596 | 2.06 |
ENSMUST00000054213.5
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chr17_-_29768531 | 2.05 |
ENSMUST00000168339.3
ENSMUST00000114683.10 ENSMUST00000234620.2 |
Tmem217
|
transmembrane protein 217 |
| chr2_+_126057020 | 2.04 |
ENSMUST00000164042.3
|
Gm17555
|
predicted gene, 17555 |
| chrX_+_110801086 | 2.01 |
ENSMUST00000207962.2
|
Gm45194
|
predicted gene 45194 |
| chr2_-_35994819 | 2.00 |
ENSMUST00000148852.4
|
Lhx6
|
LIM homeobox protein 6 |
| chr18_-_78249612 | 1.98 |
ENSMUST00000163367.3
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chrY_-_8071507 | 1.98 |
ENSMUST00000178149.3
|
Gm20825
|
predicted gene, 20825 |
| chr13_-_64460382 | 1.98 |
ENSMUST00000021938.11
ENSMUST00000221118.2 ENSMUST00000221350.2 |
Prxl2c
|
peroxiredoxin like 2C |
| chr8_+_71069476 | 1.96 |
ENSMUST00000052437.6
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr2_+_74552322 | 1.96 |
ENSMUST00000047904.4
|
Hoxd4
|
homeobox D4 |
| chr12_+_108758871 | 1.95 |
ENSMUST00000021692.9
|
Yy1
|
YY1 transcription factor |
| chr18_-_68562385 | 1.95 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr4_+_52596266 | 1.94 |
ENSMUST00000029995.6
|
Toporsl
|
topoisomerase I binding, arginine/serine-rich like |
| chr17_+_37274714 | 1.92 |
ENSMUST00000209623.2
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr16_-_23807602 | 1.91 |
ENSMUST00000023151.6
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr1_+_120530134 | 1.91 |
ENSMUST00000079721.9
|
En1
|
engrailed 1 |
| chr18_-_14105591 | 1.89 |
ENSMUST00000234025.2
|
Zfp521
|
zinc finger protein 521 |
| chr4_-_92079986 | 1.89 |
ENSMUST00000123179.2
|
Gm12666
|
predicted gene 12666 |
| chr12_+_59176543 | 1.89 |
ENSMUST00000069430.15
ENSMUST00000177370.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chrX_-_137985960 | 1.88 |
ENSMUST00000033626.15
ENSMUST00000060824.4 |
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr1_-_45542442 | 1.88 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chrY_+_24410402 | 1.87 |
ENSMUST00000187943.2
|
Gm20809
|
predicted gene, 20809 |
| chr13_+_13612136 | 1.87 |
ENSMUST00000005532.9
|
Nid1
|
nidogen 1 |
| chr7_-_104742024 | 1.86 |
ENSMUST00000042676.6
|
Olfr680-ps1
|
olfactory receptor 680, pseudogene 1 |
| chr11_-_99884818 | 1.85 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chrY_-_84576176 | 1.85 |
ENSMUST00000189463.2
|
Gm21394
|
predicted gene, 21394 |
| chr18_+_44249254 | 1.84 |
ENSMUST00000212114.2
|
Gm37797
|
predicted gene, 37797 |
| chr7_+_127311881 | 1.83 |
ENSMUST00000047393.7
|
Ctf1
|
cardiotrophin 1 |
| chr2_-_142743438 | 1.83 |
ENSMUST00000230763.2
ENSMUST00000043589.8 |
Kif16b
|
kinesin family member 16B |
| chr17_+_78815493 | 1.83 |
ENSMUST00000024880.11
ENSMUST00000232859.2 |
Vit
|
vitrin |
| chr18_+_20443795 | 1.81 |
ENSMUST00000077146.4
|
Dsg1a
|
desmoglein 1 alpha |
| chr14_+_51790247 | 1.80 |
ENSMUST00000096170.5
|
Gm5622
|
predicted gene 5622 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.3 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 4.0 | 15.9 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 3.8 | 11.3 | GO:0097037 | heme export(GO:0097037) |
| 2.9 | 11.6 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 1.6 | 4.8 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 1.5 | 4.6 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 1.3 | 8.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 1.3 | 4.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 1.3 | 7.9 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 1.3 | 5.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 1.2 | 3.7 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.2 | 3.7 | GO:2000793 | negative regulation of histone phosphorylation(GO:0033128) cell proliferation involved in heart valve development(GO:2000793) |
| 1.2 | 8.5 | GO:0046618 | drug export(GO:0046618) |
| 1.2 | 14.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 1.2 | 4.7 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 1.1 | 3.2 | GO:0033189 | response to vitamin A(GO:0033189) |
| 1.0 | 4.0 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.9 | 11.0 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.9 | 2.6 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.9 | 6.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.8 | 2.5 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.8 | 5.5 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.8 | 2.3 | GO:1903000 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.7 | 2.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.7 | 2.0 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.6 | 19.2 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.6 | 4.8 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.6 | 3.6 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.6 | 5.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.5 | 6.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.5 | 1.5 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.5 | 2.5 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.5 | 1.9 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.5 | 1.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.5 | 7.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 1.3 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.4 | 7.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.4 | 4.4 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.4 | 2.4 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.4 | 1.2 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.4 | 6.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.4 | 2.7 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.4 | 2.6 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.4 | 1.8 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 | 10.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 2.4 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.3 | 1.0 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.3 | 1.7 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.3 | 13.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 0.9 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.3 | 2.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.3 | 8.3 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.3 | 1.9 | GO:0061743 | motor learning(GO:0061743) |
| 0.3 | 1.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.3 | 1.8 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.3 | 3.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.3 | 0.8 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.2 | 2.0 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 4.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.2 | 1.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.2 | 3.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 1.0 | GO:2000293 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.2 | 1.4 | GO:1902988 | regulation of neuronal signal transduction(GO:1902847) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 0.5 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.2 | 1.4 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.2 | 0.5 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.2 | 0.7 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.2 | 0.9 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.2 | 1.3 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.2 | 1.3 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.2 | 3.1 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.2 | 4.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 0.8 | GO:0045399 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.2 | 1.7 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 4.4 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 4.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 1.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 0.5 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.2 | 1.1 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.2 | 5.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.2 | 0.5 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.2 | 6.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 0.7 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 2.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 0.8 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.2 | 1.7 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 0.6 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.1 | 0.9 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 8.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.8 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 1.4 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 2.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.7 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.8 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.4 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.1 | 2.2 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.4 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 1.4 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.4 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.4 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.1 | 2.8 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 0.7 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 3.5 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.1 | 0.9 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 7.8 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.1 | 0.3 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 0.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 1.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 2.8 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 0.8 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 1.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 5.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 1.6 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.4 | GO:0060807 | cardiogenic plate morphogenesis(GO:0003142) regulation of transcription from RNA polymerase II promoter involved in definitive endodermal cell fate specification(GO:0060807) |
| 0.1 | 3.7 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 1.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 4.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 3.2 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 4.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.3 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.2 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.1 | 4.8 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.1 | 1.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.5 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 1.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.1 | GO:0042136 | carnitine metabolic process(GO:0009437) neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.8 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.6 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 1.3 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.1 | 1.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 1.7 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 1.0 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.1 | 2.2 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.4 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.1 | 0.5 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 1.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.8 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.1 | 1.3 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.8 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 0.3 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 1.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 12.0 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 1.1 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.4 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.1 | 0.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 1.4 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.3 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 2.5 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 8.0 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.1 | 0.3 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 1.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 1.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 3.0 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.8 | GO:0090308 | maintenance of DNA methylation(GO:0010216) regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.4 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 2.9 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.5 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.8 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 2.4 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.7 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.7 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.7 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.9 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.6 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 3.1 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.8 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 2.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.2 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 8.7 | GO:0048705 | skeletal system morphogenesis(GO:0048705) |
| 0.0 | 0.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 1.1 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.4 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 2.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 33.0 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.6 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.5 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.3 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.5 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.5 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 2.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.1 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.5 | GO:0009583 | detection of light stimulus(GO:0009583) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.7 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.7 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 1.7 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 2.7 | 13.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.8 | 7.3 | GO:0008623 | CHRAC(GO:0008623) |
| 1.4 | 5.8 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 1.4 | 4.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 1.3 | 7.7 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.8 | 4.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.7 | 4.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.6 | 1.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 6.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.5 | 4.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 1.8 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.4 | 1.3 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.3 | 4.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.3 | 5.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 0.9 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 12.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 2.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 1.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.3 | 4.8 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.2 | 6.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 1.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 3.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 7.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 6.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 2.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.2 | 5.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.8 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 1.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 0.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 1.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 10.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 0.8 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.4 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 2.7 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 1.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.5 | GO:0005757 | mitochondrial permeability transition pore complex(GO:0005757) |
| 0.1 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 1.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 1.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 20.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.6 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 1.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 2.2 | GO:0043073 | male germ cell nucleus(GO:0001673) germ cell nucleus(GO:0043073) |
| 0.1 | 2.0 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 2.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 3.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 16.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.9 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 22.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 6.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 4.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 1.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.9 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 4.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 2.7 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 2.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 3.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 1.3 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 2.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 4.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 8.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 2.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 1.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 5.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.4 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 2.8 | 11.3 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 2.6 | 15.9 | GO:0032810 | sterol response element binding(GO:0032810) |
| 2.1 | 6.3 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 2.0 | 6.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 1.9 | 7.8 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 1.9 | 13.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 1.8 | 5.5 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 1.6 | 4.8 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 1.5 | 4.6 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.3 | 10.8 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 1.3 | 5.3 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 1.2 | 3.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.2 | 3.6 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 1.2 | 4.7 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 1.1 | 4.4 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.9 | 11.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.9 | 4.4 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.9 | 5.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.8 | 5.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.8 | 2.3 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.8 | 4.6 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.7 | 6.0 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.7 | 6.0 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.7 | 2.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.7 | 8.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.7 | 4.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.6 | 2.6 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.6 | 5.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 3.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.5 | 4.8 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.5 | 12.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.5 | 5.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.5 | 1.8 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.4 | 6.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.4 | 2.6 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.4 | 4.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 1.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.3 | 1.3 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.3 | 1.0 | GO:0043404 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.3 | 2.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 9.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 2.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 6.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.3 | 4.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 0.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 9.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.3 | 1.9 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.3 | 0.8 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.2 | 1.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.2 | 3.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 3.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 0.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 0.8 | GO:0030977 | taurine binding(GO:0030977) |
| 0.2 | 0.8 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.2 | 0.7 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 4.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 2.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 0.8 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 4.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 9.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 21.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 2.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 2.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.9 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 3.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.7 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 1.5 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 2.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 3.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.3 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) L-threonine ammonia-lyase activity(GO:0004794) |
| 0.1 | 3.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 6.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 2.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 2.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 8.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.7 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 2.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.7 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.3 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.1 | 1.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 2.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 1.2 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 5.6 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 2.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.5 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 1.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.3 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 1.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 3.0 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.1 | 0.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.3 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.3 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 1.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.7 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 17.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 0.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 5.5 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 1.0 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.5 | GO:0035925 | C-C chemokine binding(GO:0019957) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 8.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 1.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.3 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 19.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 7.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 10.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 22.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.8 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 14.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 4.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 7.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 12.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 13.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 9.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 6.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 18.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 5.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 12.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.0 | 13.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.6 | 11.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.5 | 6.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 4.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.3 | 4.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 9.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 7.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 4.3 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.3 | 4.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 11.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.3 | 7.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 9.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 4.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 4.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 18.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 20.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 10.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.8 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 2.6 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 1.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 2.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |