Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
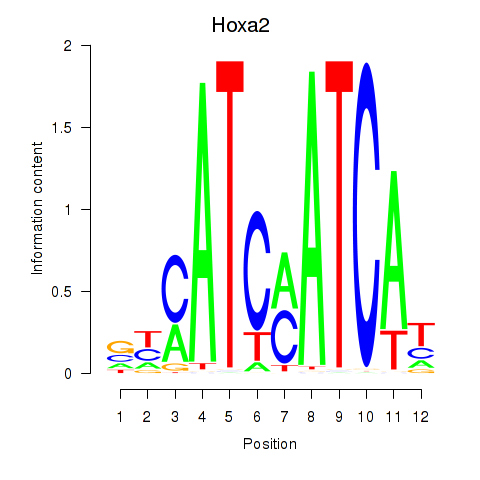
Results for Hoxa2
Z-value: 1.21
Transcription factors associated with Hoxa2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa2
|
ENSMUSG00000014704.11 | Hoxa2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa2 | mm39_v1_chr6_-_52141796_52141816 | -0.18 | 1.3e-01 | Click! |
Activity profile of Hoxa2 motif
Sorted Z-values of Hoxa2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_92659644 | 7.88 |
ENSMUST00000029527.6
|
Lce1g
|
late cornified envelope 1G |
| chr11_-_58504307 | 7.04 |
ENSMUST00000048801.8
|
Lypd8l
|
LY6/PLAUR domain containing 8 like |
| chr16_-_23709564 | 6.01 |
ENSMUST00000004480.5
|
Sst
|
somatostatin |
| chr3_+_92851790 | 5.92 |
ENSMUST00000055375.6
|
Lce3c
|
late cornified envelope 3C |
| chr3_-_92577621 | 5.62 |
ENSMUST00000029530.6
|
Lce1a2
|
late cornified envelope 1A2 |
| chr3_-_92697821 | 5.49 |
ENSMUST00000186525.2
|
Lce1j
|
late cornified envelope 1J |
| chr15_+_82136598 | 5.41 |
ENSMUST00000136948.3
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr3_+_93301003 | 5.27 |
ENSMUST00000045912.3
|
Rptn
|
repetin |
| chr17_-_31383976 | 5.26 |
ENSMUST00000235870.2
|
Tff1
|
trefoil factor 1 |
| chr1_+_106866678 | 5.15 |
ENSMUST00000112724.3
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr1_-_106980033 | 4.88 |
ENSMUST00000112717.3
|
Serpinb3a
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A |
| chr16_-_56891711 | 4.80 |
ENSMUST00000067173.8
ENSMUST00000227043.2 |
Tmem45a2
|
transmembrane protein 45A2 |
| chr3_-_92833537 | 4.70 |
ENSMUST00000067318.6
|
Lce3a
|
late cornified envelope 3A |
| chr6_+_86605146 | 4.26 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr3_-_92594516 | 4.17 |
ENSMUST00000029524.4
|
Lce1d
|
late cornified envelope 1D |
| chr7_+_30487322 | 4.16 |
ENSMUST00000189673.7
ENSMUST00000190990.7 ENSMUST00000189962.7 ENSMUST00000187493.7 ENSMUST00000098559.3 |
Krtdap
|
keratinocyte differentiation associated protein |
| chr11_-_106471420 | 4.04 |
ENSMUST00000153870.2
|
Tex2
|
testis expressed gene 2 |
| chr3_-_107851021 | 3.99 |
ENSMUST00000106684.8
ENSMUST00000106685.9 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr7_+_43441315 | 3.98 |
ENSMUST00000005891.7
|
Klk9
|
kallikrein related-peptidase 9 |
| chr7_-_44861541 | 3.97 |
ENSMUST00000033057.9
|
Dkkl1
|
dickkopf-like 1 |
| chr1_+_134038970 | 3.85 |
ENSMUST00000159963.8
ENSMUST00000160060.8 |
Chit1
|
chitinase 1 (chitotriosidase) |
| chr3_-_92627651 | 3.82 |
ENSMUST00000047153.4
|
Lce1f
|
late cornified envelope 1F |
| chr7_-_108529375 | 3.69 |
ENSMUST00000055745.5
|
Nlrp10
|
NLR family, pyrin domain containing 10 |
| chr1_+_131566223 | 3.63 |
ENSMUST00000112411.2
|
Ctse
|
cathepsin E |
| chr14_-_68893253 | 3.59 |
ENSMUST00000225767.3
ENSMUST00000111072.8 ENSMUST00000022642.6 ENSMUST00000224039.2 |
Adam28
|
a disintegrin and metallopeptidase domain 28 |
| chr1_+_107289659 | 3.54 |
ENSMUST00000027566.3
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr11_+_108271990 | 3.37 |
ENSMUST00000146050.2
ENSMUST00000152958.8 |
Apoh
|
apolipoprotein H |
| chr4_-_61972348 | 3.35 |
ENSMUST00000074018.4
|
Mup20
|
major urinary protein 20 |
| chr2_+_102536701 | 3.32 |
ENSMUST00000123759.8
ENSMUST00000005220.11 ENSMUST00000111212.8 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr18_-_32271224 | 3.32 |
ENSMUST00000234657.2
ENSMUST00000234386.2 ENSMUST00000234651.2 |
Proc
|
protein C |
| chr9_-_99599312 | 3.15 |
ENSMUST00000112882.9
ENSMUST00000131922.2 |
Cldn18
|
claudin 18 |
| chr14_+_66208498 | 3.13 |
ENSMUST00000128539.8
|
Clu
|
clusterin |
| chr9_+_98368163 | 2.97 |
ENSMUST00000189446.7
ENSMUST00000187905.7 |
Rbp2
|
retinol binding protein 2, cellular |
| chr3_-_107850707 | 2.95 |
ENSMUST00000106681.3
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr14_+_66208613 | 2.87 |
ENSMUST00000144619.2
|
Clu
|
clusterin |
| chr16_+_96162854 | 2.86 |
ENSMUST00000113794.8
|
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr10_+_128104525 | 2.80 |
ENSMUST00000050901.5
|
Apof
|
apolipoprotein F |
| chr19_-_44017637 | 2.80 |
ENSMUST00000026211.10
ENSMUST00000211830.2 |
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr1_+_131566044 | 2.79 |
ENSMUST00000073350.13
|
Ctse
|
cathepsin E |
| chr2_+_24076481 | 2.74 |
ENSMUST00000057567.3
|
Il1f9
|
interleukin 1 family, member 9 |
| chr14_-_30645503 | 2.73 |
ENSMUST00000227995.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr12_-_103979900 | 2.70 |
ENSMUST00000058464.5
|
Serpina9
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| chr7_+_26819334 | 2.67 |
ENSMUST00000003100.10
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr13_-_4329421 | 2.61 |
ENSMUST00000021632.5
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr19_+_20470056 | 2.61 |
ENSMUST00000225337.3
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr8_-_95422851 | 2.59 |
ENSMUST00000034227.6
|
Pllp
|
plasma membrane proteolipid |
| chr3_+_92864693 | 2.57 |
ENSMUST00000059053.11
|
Lce3d
|
late cornified envelope 3D |
| chr12_-_103423472 | 2.56 |
ENSMUST00000044687.7
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr12_-_103980011 | 2.54 |
ENSMUST00000164023.2
|
Serpina9
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| chr16_+_92295009 | 2.50 |
ENSMUST00000023670.4
|
Clic6
|
chloride intracellular channel 6 |
| chr19_-_4092218 | 2.41 |
ENSMUST00000237999.2
ENSMUST00000042700.12 |
Gstp2
|
glutathione S-transferase, pi 2 |
| chr7_-_28330322 | 2.41 |
ENSMUST00000040112.5
ENSMUST00000239470.2 |
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr1_-_139487951 | 2.32 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr18_-_12952925 | 2.29 |
ENSMUST00000119043.8
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr3_+_130411294 | 2.28 |
ENSMUST00000163620.8
|
Etnppl
|
ethanolamine phosphate phospholyase |
| chr9_-_70841881 | 2.27 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chr19_-_47680528 | 2.25 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr1_+_88062508 | 2.24 |
ENSMUST00000113134.8
ENSMUST00000140092.8 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr7_-_28329924 | 2.23 |
ENSMUST00000159095.2
ENSMUST00000159418.8 ENSMUST00000159560.3 |
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr13_-_42001075 | 2.15 |
ENSMUST00000179758.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr19_+_33985684 | 2.13 |
ENSMUST00000225505.2
|
Lipk
|
lipase, family member K |
| chr1_+_88030951 | 2.11 |
ENSMUST00000113135.6
ENSMUST00000113138.8 |
Ugt1a7c
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A7C UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr6_+_14901343 | 2.11 |
ENSMUST00000115477.8
|
Foxp2
|
forkhead box P2 |
| chr19_+_33985701 | 2.10 |
ENSMUST00000054260.8
|
Lipk
|
lipase, family member K |
| chr5_-_87572060 | 2.08 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr9_-_26834356 | 2.06 |
ENSMUST00000115261.3
|
Gm1110
|
predicted gene 1110 |
| chr3_+_62327089 | 2.05 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr4_+_63262775 | 2.01 |
ENSMUST00000030044.3
|
Orm1
|
orosomucoid 1 |
| chr19_+_34078333 | 2.00 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr19_+_12577317 | 2.00 |
ENSMUST00000181868.8
ENSMUST00000092931.7 |
Gm4952
|
predicted gene 4952 |
| chr13_+_23991010 | 1.97 |
ENSMUST00000006786.11
ENSMUST00000099697.3 |
Slc17a2
|
solute carrier family 17 (sodium phosphate), member 2 |
| chr6_+_41279199 | 1.97 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr3_+_28946760 | 1.96 |
ENSMUST00000099170.2
|
Gm1527
|
predicted gene 1527 |
| chr13_-_25121568 | 1.94 |
ENSMUST00000037615.7
|
Aldh5a1
|
aldhehyde dehydrogenase family 5, subfamily A1 |
| chr13_+_4109566 | 1.94 |
ENSMUST00000041768.7
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr16_+_25620652 | 1.93 |
ENSMUST00000115304.8
ENSMUST00000115305.2 ENSMUST00000040231.13 ENSMUST00000115306.8 |
Trp63
|
transformation related protein 63 |
| chr13_-_42001102 | 1.92 |
ENSMUST00000121404.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr5_-_87288177 | 1.92 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr7_+_30450896 | 1.91 |
ENSMUST00000182229.8
ENSMUST00000080518.14 ENSMUST00000182227.8 ENSMUST00000182721.8 |
Sbsn
|
suprabasin |
| chr1_-_135846858 | 1.90 |
ENSMUST00000163260.8
|
Pkp1
|
plakophilin 1 |
| chr17_+_32671689 | 1.85 |
ENSMUST00000237491.2
|
Cyp4f39
|
cytochrome P450, family 4, subfamily f, polypeptide 39 |
| chr19_+_20470114 | 1.85 |
ENSMUST00000225313.2
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr4_-_107975701 | 1.84 |
ENSMUST00000149106.8
|
Scp2
|
sterol carrier protein 2, liver |
| chr16_+_22710785 | 1.82 |
ENSMUST00000023583.7
ENSMUST00000232098.2 |
Ahsg
|
alpha-2-HS-glycoprotein |
| chr1_+_153775682 | 1.82 |
ENSMUST00000086199.12
|
Glul
|
glutamate-ammonia ligase (glutamine synthetase) |
| chr4_-_62069046 | 1.82 |
ENSMUST00000077719.4
|
Mup21
|
major urinary protein 21 |
| chr5_-_86616849 | 1.81 |
ENSMUST00000101073.3
|
Tmprss11a
|
transmembrane protease, serine 11a |
| chr3_+_134534754 | 1.77 |
ENSMUST00000029822.6
|
Tacr3
|
tachykinin receptor 3 |
| chr3_+_130411097 | 1.75 |
ENSMUST00000166187.8
ENSMUST00000072271.13 |
Etnppl
|
ethanolamine phosphate phospholyase |
| chr9_-_70842090 | 1.72 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr7_-_43155118 | 1.71 |
ENSMUST00000168883.2
|
4931406B18Rik
|
RIKEN cDNA 4931406B18 gene |
| chr1_-_121255448 | 1.70 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr4_+_98919183 | 1.61 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr18_-_56705960 | 1.60 |
ENSMUST00000174518.8
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr3_-_116762617 | 1.59 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr3_-_113368407 | 1.58 |
ENSMUST00000106540.8
|
Amy1
|
amylase 1, salivary |
| chr10_+_75768964 | 1.56 |
ENSMUST00000219839.2
|
Chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr14_-_61275340 | 1.55 |
ENSMUST00000225730.2
ENSMUST00000111236.4 |
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr18_-_56695333 | 1.54 |
ENSMUST00000066208.13
ENSMUST00000172734.8 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr1_-_121255400 | 1.53 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr12_+_57610897 | 1.53 |
ENSMUST00000101398.10
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr7_-_45019984 | 1.53 |
ENSMUST00000003971.10
|
Lin7b
|
lin-7 homolog B (C. elegans) |
| chr4_-_155095441 | 1.53 |
ENSMUST00000105631.9
ENSMUST00000139976.9 ENSMUST00000145662.9 |
Plch2
|
phospholipase C, eta 2 |
| chr2_-_134396268 | 1.52 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chrX_+_109857866 | 1.52 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr17_+_14168635 | 1.51 |
ENSMUST00000088809.6
|
Gm7168
|
predicted gene 7168 |
| chr8_-_45747883 | 1.51 |
ENSMUST00000026907.6
|
Klkb1
|
kallikrein B, plasma 1 |
| chr1_-_24139387 | 1.50 |
ENSMUST00000027337.15
|
Fam135a
|
family with sequence similarity 135, member A |
| chr17_-_15198955 | 1.48 |
ENSMUST00000231584.2
ENSMUST00000097399.6 ENSMUST00000232173.2 |
Dynlt2a3
|
dynein light chain Tctex-type 2A3 |
| chr7_+_44813363 | 1.45 |
ENSMUST00000085374.7
ENSMUST00000209634.2 |
Slc17a7
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 |
| chrX_-_142716200 | 1.45 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr10_+_33733706 | 1.44 |
ENSMUST00000218204.2
|
Sult3a1
|
sulfotransferase family 3A, member 1 |
| chr18_-_56695288 | 1.43 |
ENSMUST00000170309.8
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr1_-_135846937 | 1.43 |
ENSMUST00000027667.13
|
Pkp1
|
plakophilin 1 |
| chr2_-_5068743 | 1.43 |
ENSMUST00000027986.5
|
Optn
|
optineurin |
| chr14_+_30608478 | 1.42 |
ENSMUST00000168782.4
|
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr1_-_10038030 | 1.40 |
ENSMUST00000185184.2
|
Tcf24
|
transcription factor 24 |
| chr12_-_103956176 | 1.39 |
ENSMUST00000151709.3
ENSMUST00000176246.3 ENSMUST00000074693.13 ENSMUST00000120251.9 |
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr5_+_125552878 | 1.38 |
ENSMUST00000031445.5
|
Aacs
|
acetoacetyl-CoA synthetase |
| chrX_-_134111421 | 1.38 |
ENSMUST00000033783.2
|
Tceal6
|
transcription elongation factor A (SII)-like 6 |
| chrX_-_94209913 | 1.37 |
ENSMUST00000113873.9
ENSMUST00000113876.9 ENSMUST00000199920.5 ENSMUST00000113885.8 ENSMUST00000113883.8 ENSMUST00000196012.2 ENSMUST00000182001.8 ENSMUST00000113878.8 ENSMUST00000113882.8 ENSMUST00000182562.2 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chr5_+_67417908 | 1.36 |
ENSMUST00000037918.12
ENSMUST00000162543.8 ENSMUST00000161233.8 ENSMUST00000160352.8 |
Tmem33
|
transmembrane protein 33 |
| chr9_-_64644966 | 1.35 |
ENSMUST00000171100.2
ENSMUST00000167569.8 ENSMUST00000004892.7 ENSMUST00000172298.8 |
Rab11a
|
RAB11A, member RAS oncogene family |
| chr16_+_34605282 | 1.35 |
ENSMUST00000023538.9
|
Mylk
|
myosin, light polypeptide kinase |
| chr6_+_14901439 | 1.34 |
ENSMUST00000128567.8
|
Foxp2
|
forkhead box P2 |
| chr13_+_30843937 | 1.33 |
ENSMUST00000091672.13
|
Dusp22
|
dual specificity phosphatase 22 |
| chr6_+_54244116 | 1.33 |
ENSMUST00000114402.9
|
Chn2
|
chimerin 2 |
| chr8_-_55177510 | 1.32 |
ENSMUST00000175915.8
|
Wdr17
|
WD repeat domain 17 |
| chr17_+_32671719 | 1.32 |
ENSMUST00000236307.2
|
Cyp4f39
|
cytochrome P450, family 4, subfamily f, polypeptide 39 |
| chr19_-_8382424 | 1.32 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chrX_-_88976458 | 1.30 |
ENSMUST00000088146.3
|
4930415L06Rik
|
RIKEN cDNA 4930415L06 gene |
| chr7_-_12731594 | 1.30 |
ENSMUST00000133977.3
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr19_+_29902506 | 1.29 |
ENSMUST00000120388.9
ENSMUST00000144528.8 ENSMUST00000177518.8 |
Il33
|
interleukin 33 |
| chr2_+_129854256 | 1.29 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr2_+_121189091 | 1.28 |
ENSMUST00000000317.13
ENSMUST00000129130.3 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr17_-_47748289 | 1.28 |
ENSMUST00000061885.9
|
1700001C19Rik
|
RIKEN cDNA 1700001C19 gene |
| chr7_-_108769719 | 1.28 |
ENSMUST00000208136.2
ENSMUST00000036992.9 |
Lmo1
|
LIM domain only 1 |
| chr1_-_37996838 | 1.27 |
ENSMUST00000027254.10
ENSMUST00000114894.2 |
Lyg1
|
lysozyme G-like 1 |
| chr6_+_83133441 | 1.26 |
ENSMUST00000203203.2
|
1700003E16Rik
|
RIKEN cDNA 1700003E16 gene |
| chr7_+_43446968 | 1.26 |
ENSMUST00000085461.4
ENSMUST00000205537.2 |
Klk8
|
kallikrein related-peptidase 8 |
| chr14_+_30608433 | 1.24 |
ENSMUST00000120269.11
ENSMUST00000078490.14 ENSMUST00000006703.15 |
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr16_+_38167352 | 1.24 |
ENSMUST00000050273.9
ENSMUST00000120495.2 ENSMUST00000119704.2 |
Cox17
Gm21987
|
cytochrome c oxidase assembly protein 17, copper chaperone predicted gene 21987 |
| chr19_-_39451509 | 1.24 |
ENSMUST00000035488.3
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr16_+_13758494 | 1.23 |
ENSMUST00000141971.8
ENSMUST00000124947.8 ENSMUST00000023360.14 ENSMUST00000143697.8 |
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr2_-_162929732 | 1.23 |
ENSMUST00000094653.6
|
Gtsf1l
|
gametocyte specific factor 1-like |
| chr7_+_43418404 | 1.22 |
ENSMUST00000014063.6
|
Klk12
|
kallikrein related-peptidase 12 |
| chr3_-_58433313 | 1.21 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr1_+_87983099 | 1.21 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr4_+_146586445 | 1.20 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr6_-_21851827 | 1.20 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr16_+_78727829 | 1.19 |
ENSMUST00000114216.2
ENSMUST00000069148.13 ENSMUST00000023568.14 |
Chodl
|
chondrolectin |
| chr5_-_118382926 | 1.19 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr3_-_157630690 | 1.19 |
ENSMUST00000118539.2
|
Cth
|
cystathionase (cystathionine gamma-lyase) |
| chr10_+_29019645 | 1.18 |
ENSMUST00000092629.4
|
Soga3
|
SOGA family member 3 |
| chr3_+_92339886 | 1.17 |
ENSMUST00000067102.3
|
Sprr2k
|
small proline-rich protein 2K |
| chr10_-_75633362 | 1.17 |
ENSMUST00000120177.8
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr7_-_43155351 | 1.16 |
ENSMUST00000191516.7
ENSMUST00000013497.8 ENSMUST00000163619.8 |
4931406B18Rik
|
RIKEN cDNA 4931406B18 gene |
| chr8_-_45811774 | 1.16 |
ENSMUST00000155230.2
ENSMUST00000135912.8 |
Fam149a
|
family with sequence similarity 149, member A |
| chr17_-_53846438 | 1.15 |
ENSMUST00000056198.4
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr14_+_3576275 | 1.15 |
ENSMUST00000151926.8
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr7_+_97569156 | 1.13 |
ENSMUST00000041860.13
|
Gdpd4
|
glycerophosphodiester phosphodiesterase domain containing 4 |
| chr4_-_83203388 | 1.13 |
ENSMUST00000150522.8
|
Ttc39b
|
tetratricopeptide repeat domain 39B |
| chr4_-_115875055 | 1.13 |
ENSMUST00000049095.6
|
Faah
|
fatty acid amide hydrolase |
| chr15_-_95426419 | 1.13 |
ENSMUST00000229933.2
ENSMUST00000166170.9 |
Nell2
|
NEL-like 2 |
| chr10_-_80861239 | 1.12 |
ENSMUST00000055125.5
|
Diras1
|
DIRAS family, GTP-binding RAS-like 1 |
| chr9_+_95441652 | 1.12 |
ENSMUST00000079597.7
|
Paqr9
|
progestin and adipoQ receptor family member IX |
| chr2_-_155668567 | 1.12 |
ENSMUST00000109638.2
ENSMUST00000134278.2 |
Eif6
|
eukaryotic translation initiation factor 6 |
| chr18_-_56695259 | 1.09 |
ENSMUST00000171844.3
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr17_-_15261701 | 1.09 |
ENSMUST00000097398.12
ENSMUST00000040746.8 ENSMUST00000097400.5 |
Dynlt2a1
Dynlt2a2
|
dynein light chain Tctex-type 2A1 dynein light chain Tctex-type 2A2 |
| chr17_-_35265514 | 1.08 |
ENSMUST00000007250.14
|
Msh5
|
mutS homolog 5 |
| chr2_+_181405106 | 1.08 |
ENSMUST00000081125.11
|
Myt1
|
myelin transcription factor 1 |
| chr13_-_21726945 | 1.08 |
ENSMUST00000205976.3
ENSMUST00000175637.3 |
Olfr1366
|
olfactory receptor 1366 |
| chr1_+_165591315 | 1.08 |
ENSMUST00000111432.10
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr12_+_87073338 | 1.07 |
ENSMUST00000110187.8
ENSMUST00000156162.8 |
Tmem63c
|
transmembrane protein 63c |
| chr4_+_123077515 | 1.07 |
ENSMUST00000152194.2
|
Hpcal4
|
hippocalcin-like 4 |
| chr6_-_67743756 | 1.06 |
ENSMUST00000103306.2
|
Igkv1-131
|
immunoglobulin kappa variable 1-131 |
| chr5_-_128897086 | 1.06 |
ENSMUST00000198941.5
ENSMUST00000199537.5 |
Rimbp2
|
RIMS binding protein 2 |
| chr8_+_55053809 | 1.06 |
ENSMUST00000033917.7
|
Spata4
|
spermatogenesis associated 4 |
| chr19_-_46315543 | 1.06 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr13_-_100689212 | 1.05 |
ENSMUST00000022140.12
|
Ocln
|
occludin |
| chr2_+_25590051 | 1.04 |
ENSMUST00000077667.4
|
Obp2a
|
odorant binding protein 2A |
| chr2_+_22958179 | 1.04 |
ENSMUST00000227663.2
ENSMUST00000028121.15 ENSMUST00000227809.2 ENSMUST00000144088.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr10_-_80861357 | 1.04 |
ENSMUST00000144640.2
|
Diras1
|
DIRAS family, GTP-binding RAS-like 1 |
| chr2_+_85715984 | 1.04 |
ENSMUST00000213441.3
|
Olfr1023
|
olfactory receptor 1023 |
| chr18_-_62044871 | 1.04 |
ENSMUST00000166783.3
ENSMUST00000049378.15 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr10_-_25412010 | 1.04 |
ENSMUST00000179685.3
|
Smlr1
|
small leucine-rich protein 1 |
| chr17_+_3447465 | 1.03 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr8_-_55177352 | 1.02 |
ENSMUST00000129132.3
ENSMUST00000150488.8 ENSMUST00000127511.9 |
Wdr17
|
WD repeat domain 17 |
| chr1_-_153775944 | 1.02 |
ENSMUST00000123490.2
|
Teddm2
|
transmembrane epididymal family member 2 |
| chr13_-_100689105 | 1.02 |
ENSMUST00000159459.8
|
Ocln
|
occludin |
| chr15_+_78915071 | 1.01 |
ENSMUST00000006544.9
ENSMUST00000171999.9 |
Gcat
Gcat
|
glycine C-acetyltransferase (2-amino-3-ketobutyrate-coenzyme A ligase) glycine C-acetyltransferase (2-amino-3-ketobutyrate-coenzyme A ligase) |
| chrX_-_162810959 | 1.01 |
ENSMUST00000033739.5
|
Car5b
|
carbonic anhydrase 5b, mitochondrial |
| chr15_-_79389442 | 1.01 |
ENSMUST00000057801.8
|
Kcnj4
|
potassium inwardly-rectifying channel, subfamily J, member 4 |
| chr4_+_123077286 | 1.01 |
ENSMUST00000126995.2
|
Hpcal4
|
hippocalcin-like 4 |
| chrX_-_101950075 | 1.01 |
ENSMUST00000120808.8
ENSMUST00000121197.8 |
Dmrtc1a
|
DMRT-like family C1a |
| chr16_+_22769822 | 1.01 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr16_-_64926076 | 1.00 |
ENSMUST00000063076.6
|
Htr1f
|
5-hydroxytryptamine (serotonin) receptor 1F |
| chr8_+_3520841 | 1.00 |
ENSMUST00000047265.13
|
Tex45
|
testis expressed 45 |
| chr3_+_124114504 | 1.00 |
ENSMUST00000058994.6
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chrX_-_142716085 | 0.99 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.0 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.9 | 2.7 | GO:0018931 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.8 | 4.0 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.7 | 2.8 | GO:0032379 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.7 | 2.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.7 | 2.7 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.6 | 1.9 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) cloacal septation(GO:0060197) |
| 0.6 | 4.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.6 | 4.9 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.6 | 41.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.6 | 1.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.6 | 3.4 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.5 | 4.5 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 1.5 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.5 | 3.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.5 | 1.4 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.5 | 1.4 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.4 | 3.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.4 | 1.3 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.4 | 4.4 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.4 | 1.3 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 0.4 | 2.9 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.4 | 1.2 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.4 | 2.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.4 | 1.6 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.4 | 1.2 | GO:0042196 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.4 | 1.9 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.4 | 3.5 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 | 1.9 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.4 | 1.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.4 | 3.3 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.4 | 6.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.3 | 1.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 1.0 | GO:0097037 | heme export(GO:0097037) |
| 0.3 | 3.0 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 1.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.3 | 1.5 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.3 | 1.2 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.3 | 1.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.3 | 1.3 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.3 | 0.8 | GO:0019676 | ammonia assimilation cycle(GO:0019676) arginine biosynthetic process via ornithine(GO:0042450) |
| 0.3 | 1.6 | GO:0051005 | negative regulation of phospholipase activity(GO:0010519) negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.3 | 3.5 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.3 | 2.6 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.3 | 2.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 1.5 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 0.7 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 0.9 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.2 | 0.9 | GO:0071110 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.2 | 0.7 | GO:1901254 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.2 | 3.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.2 | 1.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.2 | 1.0 | GO:0036343 | psychomotor behavior(GO:0036343) regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.2 | 0.8 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.2 | 0.6 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.2 | 3.7 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.2 | 0.5 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.2 | 0.8 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.2 | 0.6 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.2 | 1.8 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.2 | 0.3 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 0.4 | GO:0051878 | lateral element assembly(GO:0051878) |
| 0.1 | 2.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.6 | GO:0044771 | pachytene(GO:0000239) meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) |
| 0.1 | 1.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 1.3 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 2.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.7 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 6.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 2.1 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.1 | 0.6 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.4 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.1 | 0.7 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.7 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.1 | 0.9 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.4 | GO:0021750 | cerebellar molecular layer development(GO:0021679) vestibular nucleus development(GO:0021750) |
| 0.1 | 0.3 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.1 | 0.9 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.1 | 1.9 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.4 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 2.8 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 0.8 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 1.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.3 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.1 | 0.7 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.6 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.4 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.1 | 0.3 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 1.5 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 0.2 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.5 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 1.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.4 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.6 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 1.9 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.6 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.5 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 1.5 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 0.2 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.1 | 0.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.4 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.1 | 0.6 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.1 | 0.2 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.4 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.9 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 2.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.8 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.9 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.4 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.4 | GO:0060459 | left lung development(GO:0060459) left lung morphogenesis(GO:0060460) |
| 0.1 | 0.2 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.2 | GO:0097374 | proprioception(GO:0019230) sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.6 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 0.1 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.2 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 1.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.6 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.3 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 2.6 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.5 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 2.3 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 1.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.9 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.4 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0070895 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 0.7 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 1.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 12.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 3.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.2 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.8 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.9 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.9 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 1.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 2.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.4 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.0 | 0.4 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 1.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.8 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.3 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 1.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 1.3 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.2 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 30.6 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.3 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.9 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 2.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 1.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 1.7 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 1.4 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.6 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 1.0 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 1.4 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.5 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 5.0 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 1.0 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.6 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 1.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 3.8 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 1.8 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.3 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.5 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 1.1 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.2 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.6 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.6 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 0.4 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 47.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.5 | 2.4 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.4 | 3.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 6.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.3 | 1.0 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.2 | 6.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 4.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.2 | 2.7 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 1.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 1.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 1.3 | GO:0005828 | kinetochore microtubule(GO:0005828) postsynaptic recycling endosome(GO:0098837) |
| 0.2 | 0.7 | GO:0019034 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 0.2 | 1.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 3.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.6 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.8 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 1.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 3.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.4 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.4 | GO:0000802 | transverse filament(GO:0000802) |
| 0.1 | 2.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 1.9 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.4 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.9 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 2.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.4 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.1 | 0.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 3.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 0.9 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 1.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 1.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 3.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 5.7 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.8 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 1.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 3.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 1.3 | 4.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.9 | 4.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.8 | 2.5 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.8 | 2.4 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 0.8 | 4.0 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.7 | 2.8 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.7 | 2.8 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.7 | 3.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.6 | 1.7 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.6 | 3.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.6 | 3.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 1.8 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.4 | 1.2 | GO:0047651 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.4 | 3.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.4 | 1.5 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.3 | 3.4 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.3 | 11.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.3 | 1.6 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.3 | 1.9 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.3 | 1.8 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.3 | 1.5 | GO:0015321 | sodium:inorganic phosphate symporter activity(GO:0015319) sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.3 | 2.0 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.3 | 2.4 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.3 | 10.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 0.7 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.2 | 0.9 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 0.7 | GO:0001566 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.2 | 1.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 0.9 | GO:0004080 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.2 | 1.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 1.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 1.7 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.2 | 1.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.2 | 2.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 0.8 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 3.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 1.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 4.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 1.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 2.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 3.0 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 27.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.4 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 1.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 3.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.8 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 2.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.8 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 1.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 6.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 3.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.3 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 3.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 2.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 2.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 0.4 | GO:0038100 | nodal binding(GO:0038100) |
| 0.1 | 1.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 1.9 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.7 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 0.2 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 2.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.4 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.1 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 1.0 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.4 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 0.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 0.2 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.2 | GO:0051381 | histamine binding(GO:0051381) |
| 0.1 | 1.0 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 8.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.7 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) thyroid hormone binding(GO:0070324) |
| 0.1 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 1.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 39.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 1.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 2.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.6 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 2.9 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 5.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.3 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.0 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.2 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 2.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.5 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 32.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.8 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.1 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 6.1 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.6 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 1.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.9 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 31.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 5.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 3.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 3.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 2.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 3.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 2.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 6.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 5.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 3.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 1.3 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.2 | 4.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 2.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 4.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 3.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 3.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 8.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 1.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 4.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 3.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 2.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |