Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Hoxa7_Hoxc8
Z-value: 0.82
Transcription factors associated with Hoxa7_Hoxc8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
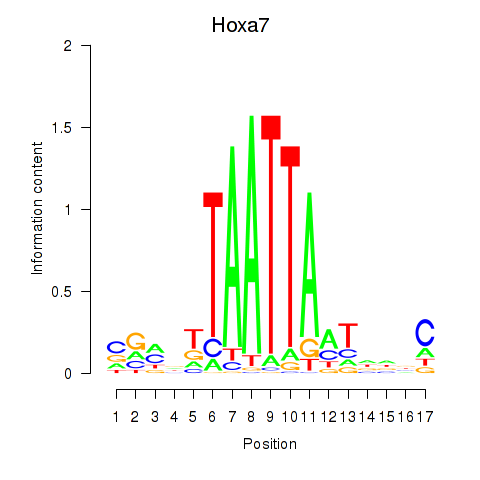
Hoxa7
|
ENSMUSG00000038236.9 | Hoxa7 |
|
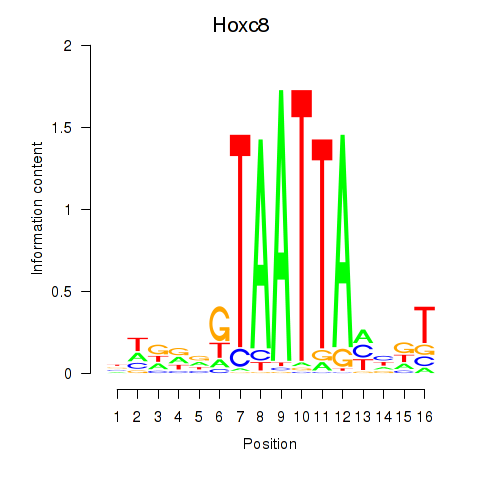
Hoxc8
|
ENSMUSG00000001657.8 | Hoxc8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc8 | mm39_v1_chr15_+_102898966_102899039 | 0.39 | 7.6e-04 | Click! |
| Hoxa7 | mm39_v1_chr6_-_52194414_52194432 | 0.17 | 1.6e-01 | Click! |
Activity profile of Hoxa7_Hoxc8 motif
Sorted Z-values of Hoxa7_Hoxc8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa7_Hoxc8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_96295011 | 11.75 |
ENSMUST00000233816.2
|
Pcp4
|
Purkinje cell protein 4 |
| chr15_-_8740218 | 10.52 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr15_-_8739893 | 9.64 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr19_+_34078333 | 8.33 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr15_-_37458768 | 6.71 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr10_+_29074950 | 5.18 |
ENSMUST00000217011.2
|
Gm49353
|
predicted gene, 49353 |
| chrX_+_9751861 | 5.18 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr6_-_137146708 | 4.49 |
ENSMUST00000117919.8
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr1_-_134883645 | 3.86 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr1_-_134883577 | 3.80 |
ENSMUST00000168381.8
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr16_-_45544960 | 3.75 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chrX_-_142716200 | 3.61 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr18_+_23886765 | 3.56 |
ENSMUST00000115830.8
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr2_+_69727599 | 3.53 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr8_+_94879235 | 3.53 |
ENSMUST00000034211.10
ENSMUST00000211930.2 ENSMUST00000211915.2 |
Mt3
|
metallothionein 3 |
| chr7_+_54485336 | 3.42 |
ENSMUST00000082373.8
|
Luzp2
|
leucine zipper protein 2 |
| chr18_+_37651393 | 3.06 |
ENSMUST00000097609.3
|
Pcdhb22
|
protocadherin beta 22 |
| chr11_+_59197746 | 2.85 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr3_+_138019040 | 2.60 |
ENSMUST00000013455.13
ENSMUST00000106247.2 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr13_+_43019718 | 2.54 |
ENSMUST00000131942.2
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr3_+_55689921 | 2.51 |
ENSMUST00000075422.6
|
Mab21l1
|
mab-21-like 1 |
| chr10_-_85847697 | 2.48 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr5_-_99091681 | 2.48 |
ENSMUST00000162619.8
ENSMUST00000162147.6 |
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr18_+_37827413 | 2.47 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr2_+_69727563 | 2.46 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_+_83554741 | 2.43 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr3_+_55369149 | 2.34 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr18_+_86729184 | 2.33 |
ENSMUST00000068423.10
|
Cbln2
|
cerebellin 2 precursor protein |
| chr8_+_46338498 | 2.28 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chrM_+_9870 | 2.27 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr14_-_109151590 | 2.26 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr2_+_124978518 | 2.25 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr19_-_50667079 | 2.25 |
ENSMUST00000209413.2
ENSMUST00000072685.13 ENSMUST00000164039.9 |
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr10_+_116013122 | 2.23 |
ENSMUST00000148731.8
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr13_+_42834989 | 2.21 |
ENSMUST00000149235.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr18_-_10706701 | 2.21 |
ENSMUST00000002549.9
ENSMUST00000117726.9 ENSMUST00000117828.9 |
Abhd3
|
abhydrolase domain containing 3 |
| chr8_+_46338557 | 2.02 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr5_-_86616849 | 2.02 |
ENSMUST00000101073.3
|
Tmprss11a
|
transmembrane protease, serine 11a |
| chr11_-_73217633 | 1.98 |
ENSMUST00000134079.2
|
Aspa
|
aspartoacylase |
| chr9_-_79885063 | 1.94 |
ENSMUST00000093811.11
|
Filip1
|
filamin A interacting protein 1 |
| chrX_+_16485937 | 1.93 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr2_+_124978612 | 1.92 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr16_-_56688024 | 1.89 |
ENSMUST00000232373.2
|
Tmem45a
|
transmembrane protein 45a |
| chr9_-_79884920 | 1.86 |
ENSMUST00000239133.2
|
Filip1
|
filamin A interacting protein 1 |
| chr4_-_114991478 | 1.85 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr10_+_127226180 | 1.82 |
ENSMUST00000077046.12
ENSMUST00000105250.9 |
R3hdm2
|
R3H domain containing 2 |
| chr13_-_53627110 | 1.79 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr3_-_50398027 | 1.74 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr4_+_108576846 | 1.74 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr5_+_29400981 | 1.73 |
ENSMUST00000160888.8
ENSMUST00000159272.8 ENSMUST00000001247.12 ENSMUST00000161398.8 ENSMUST00000160246.8 |
Rnf32
|
ring finger protein 32 |
| chr2_+_32496990 | 1.72 |
ENSMUST00000095045.9
ENSMUST00000095044.10 ENSMUST00000126636.8 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr9_+_113641615 | 1.69 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr9_-_119897328 | 1.67 |
ENSMUST00000177637.2
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chrX_+_113384297 | 1.65 |
ENSMUST00000133447.2
|
Klhl4
|
kelch-like 4 |
| chr2_+_110551927 | 1.63 |
ENSMUST00000111017.9
|
Muc15
|
mucin 15 |
| chr10_-_20600442 | 1.61 |
ENSMUST00000170265.8
|
Pde7b
|
phosphodiesterase 7B |
| chr2_+_32496957 | 1.56 |
ENSMUST00000113290.8
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr4_-_114991174 | 1.55 |
ENSMUST00000051400.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr2_+_110551685 | 1.45 |
ENSMUST00000111016.9
|
Muc15
|
mucin 15 |
| chrX_-_98514278 | 1.44 |
ENSMUST00000113797.4
ENSMUST00000113790.8 ENSMUST00000036354.7 ENSMUST00000167246.2 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chr10_+_115854118 | 1.43 |
ENSMUST00000063470.11
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr9_+_109881083 | 1.39 |
ENSMUST00000164930.8
ENSMUST00000199498.5 |
Map4
|
microtubule-associated protein 4 |
| chr16_+_13176238 | 1.37 |
ENSMUST00000149359.2
|
Mrtfb
|
myocardin related transcription factor B |
| chr11_-_73217298 | 1.36 |
ENSMUST00000155630.9
|
Aspa
|
aspartoacylase |
| chr9_-_119897358 | 1.35 |
ENSMUST00000064165.5
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr15_+_21111428 | 1.34 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chrX_-_100129626 | 1.34 |
ENSMUST00000113710.8
|
Slc7a3
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 |
| chr19_+_5524701 | 1.33 |
ENSMUST00000165485.8
ENSMUST00000166253.8 ENSMUST00000167371.8 ENSMUST00000167855.8 ENSMUST00000070118.14 |
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr13_-_54713180 | 1.23 |
ENSMUST00000026989.15
|
4833439L19Rik
|
RIKEN cDNA 4833439L19 gene |
| chr2_+_83554770 | 1.20 |
ENSMUST00000141725.3
|
Itgav
|
integrin alpha V |
| chr12_-_40298072 | 1.15 |
ENSMUST00000169926.8
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr10_+_97318223 | 1.14 |
ENSMUST00000163448.4
|
Dcn
|
decorin |
| chr5_+_104350475 | 1.08 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr16_-_88303859 | 1.02 |
ENSMUST00000069549.3
|
Cldn17
|
claudin 17 |
| chr2_+_110427643 | 1.00 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chrX_+_151909893 | 0.98 |
ENSMUST00000163801.2
|
Foxr2
|
forkhead box R2 |
| chr19_-_32173824 | 0.90 |
ENSMUST00000151822.2
|
Sgms1
|
sphingomyelin synthase 1 |
| chr14_-_36641270 | 0.87 |
ENSMUST00000182797.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr6_-_93769426 | 0.86 |
ENSMUST00000204788.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chrM_+_14138 | 0.83 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr16_-_19241884 | 0.78 |
ENSMUST00000206110.4
|
Olfr165
|
olfactory receptor 165 |
| chr7_-_25112256 | 0.77 |
ENSMUST00000200880.4
ENSMUST00000074040.4 |
Cxcl17
|
chemokine (C-X-C motif) ligand 17 |
| chr19_-_38807600 | 0.76 |
ENSMUST00000025963.8
|
Noc3l
|
NOC3 like DNA replication regulator |
| chr4_-_14621805 | 0.74 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr18_+_31742565 | 0.69 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr9_+_38119661 | 0.69 |
ENSMUST00000211975.3
|
Olfr893
|
olfactory receptor 893 |
| chr7_-_103113358 | 0.69 |
ENSMUST00000214347.2
|
Olfr607
|
olfactory receptor 607 |
| chr2_+_22959452 | 0.68 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr17_+_24072493 | 0.68 |
ENSMUST00000061725.8
|
Prss32
|
protease, serine 32 |
| chr3_-_72875187 | 0.65 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr9_+_20148415 | 0.64 |
ENSMUST00000086474.6
|
Olfr872
|
olfactory receptor 872 |
| chrX_+_164953444 | 0.63 |
ENSMUST00000130880.9
ENSMUST00000056410.11 ENSMUST00000096252.10 ENSMUST00000087169.11 |
Gemin8
|
gem nuclear organelle associated protein 8 |
| chr8_+_23901506 | 0.62 |
ENSMUST00000033952.8
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr19_+_41921903 | 0.61 |
ENSMUST00000224258.2
ENSMUST00000026154.9 ENSMUST00000224896.2 |
Zdhhc16
|
zinc finger, DHHC domain containing 16 |
| chr8_-_49008305 | 0.59 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chrM_+_9459 | 0.59 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr2_+_110551976 | 0.58 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
| chr14_-_14255736 | 0.56 |
ENSMUST00000170111.3
|
Kctd6
|
potassium channel tetramerisation domain containing 6 |
| chrX_+_158086253 | 0.55 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr3_+_5283577 | 0.54 |
ENSMUST00000175866.8
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr15_+_25774070 | 0.51 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr19_+_53933271 | 0.51 |
ENSMUST00000025932.9
|
Shoc2
|
Shoc2, leucine rich repeat scaffold protein |
| chr3_+_5283606 | 0.50 |
ENSMUST00000026284.13
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr7_-_107696793 | 0.49 |
ENSMUST00000217304.2
|
Olfr482
|
olfactory receptor 482 |
| chr19_-_53933052 | 0.48 |
ENSMUST00000135402.4
|
Bbip1
|
BBSome interacting protein 1 |
| chr11_+_102175985 | 0.44 |
ENSMUST00000156326.2
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chrM_-_14061 | 0.44 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr5_-_87716882 | 0.42 |
ENSMUST00000113314.3
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr10_-_43934774 | 0.42 |
ENSMUST00000239010.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr1_-_4563821 | 0.42 |
ENSMUST00000191939.2
|
Sox17
|
SRY (sex determining region Y)-box 17 |
| chr13_-_103042554 | 0.42 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr14_+_80237691 | 0.40 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr9_+_38883609 | 0.40 |
ENSMUST00000216238.3
|
Olfr933
|
olfactory receptor 933 |
| chr12_+_87840841 | 0.39 |
ENSMUST00000110147.3
|
Eif1ad3
|
eukaryotic translation initiation factor 1A domain containing 3 |
| chr11_+_73051228 | 0.38 |
ENSMUST00000006104.10
ENSMUST00000135202.8 ENSMUST00000136894.3 |
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr5_-_66672158 | 0.38 |
ENSMUST00000161879.8
ENSMUST00000159357.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr3_-_59127571 | 0.38 |
ENSMUST00000199675.2
ENSMUST00000170388.6 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr15_-_75777210 | 0.36 |
ENSMUST00000127550.2
|
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr6_+_132572871 | 0.36 |
ENSMUST00000076061.4
ENSMUST00000178961.2 |
Prp2
|
proline rich protein 2 |
| chr11_+_96177449 | 0.36 |
ENSMUST00000049352.8
|
Hoxb7
|
homeobox B7 |
| chr10_-_75946790 | 0.35 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr7_-_5325456 | 0.33 |
ENSMUST00000207520.2
|
Nlrp2
|
NLR family, pyrin domain containing 2 |
| chrX_+_96116202 | 0.32 |
ENSMUST00000033556.4
|
Pgr15l
|
G protein-coupled receptor 15-like |
| chr4_-_154721288 | 0.31 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr2_+_83554868 | 0.30 |
ENSMUST00000111740.9
|
Itgav
|
integrin alpha V |
| chr2_-_88559941 | 0.29 |
ENSMUST00000099815.2
|
Olfr1197
|
olfactory receptor 1197 |
| chr11_+_73489420 | 0.25 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chr2_+_87854404 | 0.25 |
ENSMUST00000217006.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr10_+_102376109 | 0.23 |
ENSMUST00000055355.6
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr18_+_32200781 | 0.22 |
ENSMUST00000025243.5
ENSMUST00000212675.2 |
Iws1
|
IWS1, SUPT6 interacting protein |
| chr9_+_21634779 | 0.20 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr13_+_22534534 | 0.18 |
ENSMUST00000226909.2
ENSMUST00000227167.2 ENSMUST00000226786.2 |
Vmn1r198
|
vomeronasal 1 receptor 198 |
| chr19_+_13339600 | 0.17 |
ENSMUST00000215096.2
|
Olfr1467
|
olfactory receptor 1467 |
| chr11_+_29323618 | 0.17 |
ENSMUST00000040182.13
ENSMUST00000109477.2 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr2_+_22959223 | 0.17 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr16_+_58967409 | 0.16 |
ENSMUST00000216957.3
|
Olfr195
|
olfactory receptor 195 |
| chr5_+_67125759 | 0.16 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chr10_+_75983285 | 0.15 |
ENSMUST00000020450.4
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr17_+_79919267 | 0.14 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr2_-_85481048 | 0.13 |
ENSMUST00000215548.3
|
Olfr1002
|
olfactory receptor 1002 |
| chr13_-_23041731 | 0.13 |
ENSMUST00000228645.2
|
Vmn1r211
|
vomeronasal 1 receptor 211 |
| chr15_-_100321973 | 0.12 |
ENSMUST00000154676.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr3_-_86455575 | 0.11 |
ENSMUST00000077524.4
|
Mab21l2
|
mab-21-like 2 |
| chr2_-_88994435 | 0.11 |
ENSMUST00000099793.3
|
Olfr1224-ps1
|
olfactory receptor 1224, pseudogene 1 |
| chr18_+_4920513 | 0.11 |
ENSMUST00000126977.8
|
Svil
|
supervillin |
| chr6_+_30541581 | 0.09 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr2_-_86109346 | 0.08 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chrX_-_142716085 | 0.08 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr7_-_102638531 | 0.06 |
ENSMUST00000215606.2
|
Olfr578
|
olfactory receptor 578 |
| chr6_-_122317484 | 0.05 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr3_+_103739877 | 0.05 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chr19_-_41921676 | 0.03 |
ENSMUST00000075280.12
ENSMUST00000112123.4 |
Exosc1
|
exosome component 1 |
| chr2_-_87570322 | 0.03 |
ENSMUST00000214573.2
|
Olfr1138
|
olfactory receptor 1138 |
| chr8_-_19041062 | 0.02 |
ENSMUST00000066282.4
|
Defb37
|
defensin beta 37 |
| chr16_+_35803794 | 0.01 |
ENSMUST00000173555.8
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr1_+_87983189 | 0.01 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr16_-_93726399 | 0.00 |
ENSMUST00000177648.8
ENSMUST00000142083.2 |
Cldn14
|
claudin 14 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 20.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.5 | 6.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 1.3 | 3.9 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 1.2 | 3.5 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 1.0 | 3.0 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.7 | 11.7 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.6 | 2.5 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.6 | 3.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.6 | 1.8 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.5 | 3.3 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.3 | 1.9 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.3 | 3.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 3.1 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.2 | 1.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.2 | 0.6 | GO:2000041 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.2 | 3.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 1.7 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.4 | GO:0060807 | cardiogenic plate morphogenesis(GO:0003142) regulation of transcription from RNA polymerase II promoter involved in definitive endodermal cell fate specification(GO:0060807) regulation of cardiac cell fate specification(GO:2000043) |
| 0.1 | 0.8 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 5.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 2.8 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.4 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.8 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 1.2 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.2 | GO:1903978 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) regulation of microglial cell activation(GO:1903978) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 2.2 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 6.7 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.4 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 0.5 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 2.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.8 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 1.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 4.8 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 4.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 3.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.2 | GO:0010793 | regulation of histone H3-K36 methylation(GO:0000414) regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.4 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 4.9 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.1 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.0 | 0.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 4.8 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.6 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.3 | GO:0050718 | positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0034683 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.8 | 11.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.6 | 1.7 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 7.7 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 3.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.8 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 0.2 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 19.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 3.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 2.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.6 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.1 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 8.7 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 4.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 1.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 4.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 7.0 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 20.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.1 | 3.3 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.8 | 6.9 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.4 | 2.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.4 | 2.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.3 | 3.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 1.7 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.3 | 1.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 1.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 1.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.6 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 0.9 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 6.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 5.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 3.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 0.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 4.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 3.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 5.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 10.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 2.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 9.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 3.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0015100 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 3.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 5.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.9 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 3.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 3.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 23.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 3.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 2.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 4.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.0 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |