Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
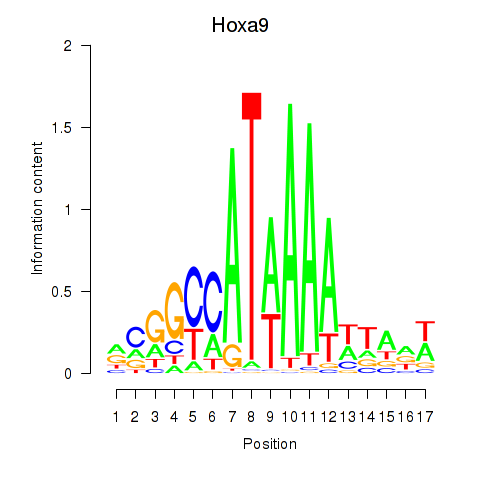
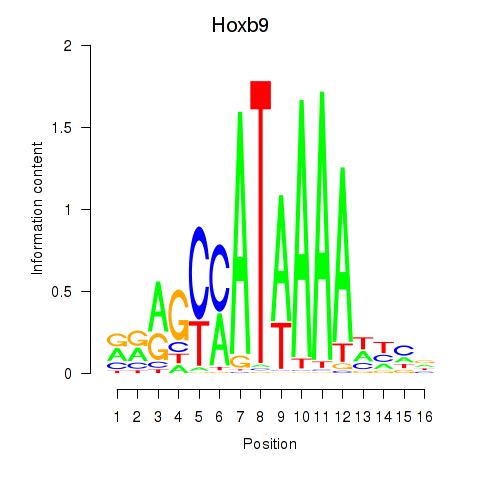
Results for Hoxa9_Hoxb9
Z-value: 0.94
Transcription factors associated with Hoxa9_Hoxb9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa9
|
ENSMUSG00000038227.16 | Hoxa9 |
|
Hoxb9
|
ENSMUSG00000020875.10 | Hoxb9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa9 | mm39_v1_chr6_-_52203146_52203176 | -0.32 | 5.8e-03 | Click! |
| Hoxb9 | mm39_v1_chr11_+_96162283_96162349 | -0.21 | 7.0e-02 | Click! |
Activity profile of Hoxa9_Hoxb9 motif
Sorted Z-values of Hoxa9_Hoxb9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa9_Hoxb9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_110781268 | 6.73 |
ENSMUST00000099623.10
|
Ano3
|
anoctamin 3 |
| chr19_-_45800730 | 5.29 |
ENSMUST00000086993.11
|
Kcnip2
|
Kv channel-interacting protein 2 |
| chr11_-_79394904 | 4.97 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr18_+_20443795 | 3.44 |
ENSMUST00000077146.4
|
Dsg1a
|
desmoglein 1 alpha |
| chr2_-_17395765 | 3.26 |
ENSMUST00000177966.2
|
Nebl
|
nebulette |
| chr3_+_138019040 | 3.24 |
ENSMUST00000013455.13
ENSMUST00000106247.2 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr11_-_42070517 | 2.99 |
ENSMUST00000206105.2
ENSMUST00000153147.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr19_-_50667079 | 2.98 |
ENSMUST00000209413.2
ENSMUST00000072685.13 ENSMUST00000164039.9 |
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr14_-_57371041 | 2.73 |
ENSMUST00000039380.9
|
Gjb6
|
gap junction protein, beta 6 |
| chr2_-_121381832 | 2.59 |
ENSMUST00000212518.2
|
Frmd5
|
FERM domain containing 5 |
| chr18_-_31450095 | 2.57 |
ENSMUST00000139924.2
ENSMUST00000153060.8 |
Rit2
|
Ras-like without CAAX 2 |
| chr2_+_22512195 | 2.56 |
ENSMUST00000028123.4
|
Gad2
|
glutamic acid decarboxylase 2 |
| chr2_-_57942844 | 2.53 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein |
| chr1_+_107439145 | 2.51 |
ENSMUST00000009356.11
ENSMUST00000064916.9 |
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr7_-_142215595 | 2.36 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr4_-_49549489 | 2.14 |
ENSMUST00000029987.10
|
Aldob
|
aldolase B, fructose-bisphosphate |
| chr7_+_43339842 | 2.13 |
ENSMUST00000056329.7
|
Klk14
|
kallikrein related-peptidase 14 |
| chr2_+_124978518 | 2.08 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr16_+_91022300 | 1.91 |
ENSMUST00000035608.10
|
Olig2
|
oligodendrocyte transcription factor 2 |
| chr2_+_124978612 | 1.89 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr2_+_58645189 | 1.85 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr5_+_128677863 | 1.83 |
ENSMUST00000117102.4
|
Fzd10
|
frizzled class receptor 10 |
| chr18_+_20509771 | 1.81 |
ENSMUST00000076737.7
|
Dsg1b
|
desmoglein 1 beta |
| chr1_+_66426127 | 1.80 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr15_-_101482320 | 1.71 |
ENSMUST00000042957.6
|
Krt75
|
keratin 75 |
| chr1_+_66403774 | 1.66 |
ENSMUST00000077355.12
ENSMUST00000114012.8 |
Map2
|
microtubule-associated protein 2 |
| chr3_-_26386609 | 1.62 |
ENSMUST00000193603.6
|
Nlgn1
|
neuroligin 1 |
| chr15_+_81820954 | 1.53 |
ENSMUST00000038757.8
ENSMUST00000230633.2 |
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr8_+_94537910 | 1.52 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr11_-_99134885 | 1.50 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr17_-_91400142 | 1.47 |
ENSMUST00000160800.9
|
Nrxn1
|
neurexin I |
| chr18_+_20380397 | 1.41 |
ENSMUST00000054128.7
|
Dsg1c
|
desmoglein 1 gamma |
| chr6_-_24168082 | 1.38 |
ENSMUST00000031713.9
|
Slc13a1
|
solute carrier family 13 (sodium/sulfate symporters), member 1 |
| chr18_-_31580436 | 1.38 |
ENSMUST00000025110.5
|
Syt4
|
synaptotagmin IV |
| chr6_+_108771840 | 1.36 |
ENSMUST00000204483.2
|
Arl8b
|
ADP-ribosylation factor-like 8B |
| chr18_+_12732951 | 1.35 |
ENSMUST00000234255.2
ENSMUST00000169401.8 |
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chrX_-_144131890 | 1.33 |
ENSMUST00000040084.10
ENSMUST00000123443.2 |
Lhfpl1
|
lipoma HMGIC fusion partner-like 1 |
| chr7_-_28330322 | 1.33 |
ENSMUST00000040112.5
ENSMUST00000239470.2 |
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr10_-_116732813 | 1.28 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr19_-_57185861 | 1.28 |
ENSMUST00000111550.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr7_-_28329924 | 1.28 |
ENSMUST00000159095.2
ENSMUST00000159418.8 ENSMUST00000159560.3 |
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr2_-_32977182 | 1.26 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr2_-_32976378 | 1.23 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr8_+_113296675 | 1.22 |
ENSMUST00000034225.7
ENSMUST00000118171.8 |
Cntnap4
|
contactin associated protein-like 4 |
| chr2_+_58644922 | 1.18 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr2_-_86180622 | 1.17 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chrX_+_136552469 | 1.13 |
ENSMUST00000075471.4
|
Il1rapl2
|
interleukin 1 receptor accessory protein-like 2 |
| chr7_+_28393419 | 1.12 |
ENSMUST00000108280.2
|
Fbxo27
|
F-box protein 27 |
| chr19_-_7319211 | 1.12 |
ENSMUST00000171393.8
|
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr14_+_20724378 | 1.11 |
ENSMUST00000224492.2
ENSMUST00000223751.2 ENSMUST00000225108.2 ENSMUST00000224754.2 |
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr5_+_145141333 | 1.07 |
ENSMUST00000085671.10
ENSMUST00000031601.8 |
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr6_-_139478919 | 1.06 |
ENSMUST00000170650.3
|
Rergl
|
RERG/RAS-like |
| chrX_-_74621828 | 1.06 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr14_+_20724366 | 1.04 |
ENSMUST00000048657.10
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr3_+_108498595 | 1.04 |
ENSMUST00000051145.15
|
Wdr47
|
WD repeat domain 47 |
| chr6_-_87958611 | 1.03 |
ENSMUST00000056403.7
|
H1f10
|
H1.10 linker histone |
| chr7_-_101519973 | 1.02 |
ENSMUST00000126204.8
ENSMUST00000155311.2 ENSMUST00000106983.8 ENSMUST00000123630.8 |
Folr1
|
folate receptor 1 (adult) |
| chr6_+_104469751 | 1.00 |
ENSMUST00000161446.2
ENSMUST00000161070.8 ENSMUST00000089215.12 |
Cntn6
|
contactin 6 |
| chr3_+_82933383 | 1.00 |
ENSMUST00000029630.15
ENSMUST00000166581.4 |
Fga
|
fibrinogen alpha chain |
| chr3_+_93052089 | 0.99 |
ENSMUST00000107300.7
ENSMUST00000195515.2 |
Crnn
|
cornulin |
| chr7_-_101513300 | 0.99 |
ENSMUST00000106981.8
|
Folr1
|
folate receptor 1 (adult) |
| chr18_+_38809771 | 0.94 |
ENSMUST00000134388.2
ENSMUST00000148850.8 |
9630014M24Rik
Arhgap26
|
RIKEN cDNA 9630014M24 gene Rho GTPase activating protein 26 |
| chr11_-_78277384 | 0.91 |
ENSMUST00000108294.2
|
Foxn1
|
forkhead box N1 |
| chr5_-_77243123 | 0.90 |
ENSMUST00000113453.9
|
Hopx
|
HOP homeobox |
| chr2_+_15531281 | 0.90 |
ENSMUST00000146205.3
|
Malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr13_-_59890564 | 0.88 |
ENSMUST00000055343.3
|
1700014D04Rik
|
RIKEN cDNA 1700014D04 gene |
| chr9_+_43222104 | 0.87 |
ENSMUST00000034511.7
|
Trim29
|
tripartite motif-containing 29 |
| chr5_-_77243072 | 0.86 |
ENSMUST00000120827.9
|
Hopx
|
HOP homeobox |
| chr14_-_31503869 | 0.84 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr3_+_84859453 | 0.83 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr7_-_101519872 | 0.82 |
ENSMUST00000106986.9
|
Folr1
|
folate receptor 1 (adult) |
| chrX_-_100128958 | 0.80 |
ENSMUST00000101362.8
ENSMUST00000073927.5 |
Slc7a3
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 |
| chr8_+_67933573 | 0.78 |
ENSMUST00000212171.2
|
Nat1
|
N-acetyl transferase 1 |
| chr15_+_6552270 | 0.78 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein |
| chr10_-_80426125 | 0.77 |
ENSMUST00000187646.2
ENSMUST00000191440.7 ENSMUST00000003436.12 |
Abhd17a
|
abhydrolase domain containing 17A |
| chr18_-_44292952 | 0.77 |
ENSMUST00000181652.9
|
Gm10267
|
predicted gene 10267 |
| chr19_-_7319157 | 0.76 |
ENSMUST00000164205.8
ENSMUST00000165286.8 ENSMUST00000168324.2 ENSMUST00000032557.15 |
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr9_+_64142483 | 0.74 |
ENSMUST00000039011.4
|
Uchl4
|
ubiquitin carboxyl-terminal esterase L4 |
| chr6_-_122317484 | 0.74 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr4_+_44012638 | 0.72 |
ENSMUST00000107847.10
ENSMUST00000170241.8 ENSMUST00000107849.10 ENSMUST00000107851.10 ENSMUST00000107845.4 |
Clta
|
clathrin, light polypeptide (Lca) |
| chr19_-_57185928 | 0.71 |
ENSMUST00000111544.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr7_-_101519914 | 0.70 |
ENSMUST00000106985.8
ENSMUST00000151706.8 |
Folr1
|
folate receptor 1 (adult) |
| chr14_-_70561231 | 0.67 |
ENSMUST00000151011.8
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr7_-_101519791 | 0.67 |
ENSMUST00000124026.8
|
Folr1
|
folate receptor 1 (adult) |
| chr14_+_56255422 | 0.67 |
ENSMUST00000022836.6
|
Mcpt1
|
mast cell protease 1 |
| chr3_+_66893280 | 0.65 |
ENSMUST00000161726.2
ENSMUST00000160504.2 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr15_+_81821112 | 0.63 |
ENSMUST00000135663.2
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr7_-_135130374 | 0.63 |
ENSMUST00000053716.8
|
Clrn3
|
clarin 3 |
| chr7_-_85985625 | 0.62 |
ENSMUST00000069279.5
|
Olfr307
|
olfactory receptor 307 |
| chr6_+_134958681 | 0.62 |
ENSMUST00000167323.3
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr19_-_57185988 | 0.61 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr2_-_23938869 | 0.61 |
ENSMUST00000114497.2
|
Hnmt
|
histamine N-methyltransferase |
| chr8_-_5155347 | 0.60 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr3_-_96812610 | 0.58 |
ENSMUST00000029738.14
|
Gpr89
|
G protein-coupled receptor 89 |
| chr13_-_21826688 | 0.58 |
ENSMUST00000205631.3
|
Olfr11
|
olfactory receptor 11 |
| chr8_+_27532623 | 0.57 |
ENSMUST00000209856.2
ENSMUST00000098851.12 ENSMUST00000211393.2 ENSMUST00000211518.2 |
Plpbp
|
pyridoxal phosphate binding protein |
| chr4_-_49597837 | 0.57 |
ENSMUST00000042750.3
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr18_+_37898633 | 0.56 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr1_+_177557380 | 0.55 |
ENSMUST00000016106.6
|
1700016C15Rik
|
RIKEN cDNA 1700016C15 gene |
| chr1_+_13738967 | 0.54 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr7_+_55699883 | 0.53 |
ENSMUST00000205653.2
ENSMUST00000076226.13 |
Herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr13_-_48746836 | 0.53 |
ENSMUST00000238995.2
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr7_+_27977141 | 0.52 |
ENSMUST00000094651.4
|
Eid2b
|
EP300 interacting inhibitor of differentiation 2B |
| chr8_-_55340024 | 0.51 |
ENSMUST00000176866.8
|
Wdr17
|
WD repeat domain 17 |
| chr5_-_73349191 | 0.49 |
ENSMUST00000176910.3
|
Fryl
|
FRY like transcription coactivator |
| chr11_+_66969119 | 0.49 |
ENSMUST00000108689.8
ENSMUST00000007301.14 ENSMUST00000165221.2 |
Myh3
|
myosin, heavy polypeptide 3, skeletal muscle, embryonic |
| chr11_-_109188947 | 0.48 |
ENSMUST00000020920.10
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr6_+_146944253 | 0.47 |
ENSMUST00000036111.10
|
Mrps35
|
mitochondrial ribosomal protein S35 |
| chr2_-_80277969 | 0.47 |
ENSMUST00000028389.4
|
Frzb
|
frizzled-related protein |
| chr5_+_137786031 | 0.47 |
ENSMUST00000035852.14
|
Zcwpw1
|
zinc finger, CW type with PWWP domain 1 |
| chr3_-_95014218 | 0.46 |
ENSMUST00000107233.9
|
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha |
| chr4_-_88562696 | 0.46 |
ENSMUST00000105149.3
|
Ifna13
|
interferon alpha 13 |
| chr16_-_17348882 | 0.45 |
ENSMUST00000231548.2
ENSMUST00000232041.2 ENSMUST00000231288.2 |
Thap7
|
THAP domain containing 7 |
| chr6_+_51500881 | 0.45 |
ENSMUST00000049152.15
|
Snx10
|
sorting nexin 10 |
| chr6_+_132928065 | 0.44 |
ENSMUST00000070991.5
|
Tas2r129
|
taste receptor, type 2, member 129 |
| chr16_+_26281885 | 0.43 |
ENSMUST00000161053.8
ENSMUST00000115302.2 |
Cldn16
|
claudin 16 |
| chr14_+_54413727 | 0.42 |
ENSMUST00000103701.2
|
Traj41
|
T cell receptor alpha joining 41 |
| chr4_-_119217079 | 0.42 |
ENSMUST00000143494.3
ENSMUST00000154606.9 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr14_+_56279498 | 0.41 |
ENSMUST00000015576.6
|
Mcpt2
|
mast cell protease 2 |
| chr3_+_98187743 | 0.41 |
ENSMUST00000120541.8
ENSMUST00000090746.3 |
Hmgcs2
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 |
| chr9_-_60594742 | 0.39 |
ENSMUST00000114032.8
ENSMUST00000166168.8 ENSMUST00000132366.2 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr5_-_32903687 | 0.39 |
ENSMUST00000135248.2
|
Pisd
|
phosphatidylserine decarboxylase |
| chr7_-_106423873 | 0.38 |
ENSMUST00000208895.3
|
Olfr702
|
olfactory receptor 702 |
| chr10_+_3822667 | 0.38 |
ENSMUST00000136671.8
ENSMUST00000042438.13 |
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr10_-_130265572 | 0.37 |
ENSMUST00000171811.4
|
Vmn2r85
|
vomeronasal 2, receptor 85 |
| chr9_+_38119661 | 0.37 |
ENSMUST00000211975.3
|
Olfr893
|
olfactory receptor 893 |
| chr8_+_27532583 | 0.36 |
ENSMUST00000033875.10
ENSMUST00000209525.2 |
Plpbp
|
pyridoxal phosphate binding protein |
| chr12_-_81615248 | 0.36 |
ENSMUST00000008582.4
|
Adam21
|
a disintegrin and metallopeptidase domain 21 |
| chr13_-_21765822 | 0.35 |
ENSMUST00000206526.3
ENSMUST00000213912.2 |
Olfr1364
|
olfactory receptor 1364 |
| chr11_-_109188892 | 0.34 |
ENSMUST00000106706.8
|
Rgs9
|
regulator of G-protein signaling 9 |
| chrX_+_71025128 | 0.34 |
ENSMUST00000114569.2
|
Fate1
|
fetal and adult testis expressed 1 |
| chr2_-_165129675 | 0.34 |
ENSMUST00000109299.8
ENSMUST00000109298.8 ENSMUST00000130393.2 ENSMUST00000017808.14 ENSMUST00000131409.2 ENSMUST00000156134.8 ENSMUST00000133961.8 |
Slc35c2
|
solute carrier family 35, member C2 |
| chr3_+_118355778 | 0.33 |
ENSMUST00000039177.12
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr2_+_147206910 | 0.32 |
ENSMUST00000109968.3
|
Pax1
|
paired box 1 |
| chr8_-_25164767 | 0.32 |
ENSMUST00000033957.12
|
Adam18
|
a disintegrin and metallopeptidase domain 18 |
| chr3_+_66893031 | 0.31 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr10_+_129153986 | 0.31 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr7_+_107501834 | 0.30 |
ENSMUST00000210420.2
|
Olfr472
|
olfactory receptor 472 |
| chr9_-_101128976 | 0.30 |
ENSMUST00000075941.12
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr16_-_17348996 | 0.30 |
ENSMUST00000100125.12
|
Thap7
|
THAP domain containing 7 |
| chr9_-_19795639 | 0.29 |
ENSMUST00000073765.6
|
Olfr862
|
olfactory receptor 862 |
| chr2_+_88479038 | 0.29 |
ENSMUST00000120518.4
|
Olfr1192-ps1
|
olfactory receptor 1192, pseudogene 1 |
| chr11_-_99941369 | 0.28 |
ENSMUST00000007318.2
|
Krt31
|
keratin 31 |
| chr3_+_66892979 | 0.28 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr6_+_70565173 | 0.27 |
ENSMUST00000103398.2
|
Igkv3-9
|
immunoglobulin kappa variable 3-9 |
| chr19_-_10995390 | 0.27 |
ENSMUST00000177684.3
|
Ms4a18
|
membrane-spanning 4-domains, subfamily A, member 18 |
| chr9_+_103917821 | 0.27 |
ENSMUST00000216593.2
ENSMUST00000147249.3 |
Nphp3
Gm28305
|
nephronophthisis 3 (adolescent) predicted gene 28305 |
| chrX_+_158623460 | 0.27 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr9_+_44966464 | 0.26 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chr1_-_107011236 | 0.26 |
ENSMUST00000023861.5
|
Serpinb3d
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3D |
| chr11_+_115294560 | 0.26 |
ENSMUST00000153983.8
ENSMUST00000106539.10 ENSMUST00000103036.5 |
Mrpl58
|
mitochondrial ribosomal protein L58 |
| chr7_-_29643172 | 0.26 |
ENSMUST00000108212.8
|
Zfp74
|
zinc finger protein 74 |
| chr7_-_12468931 | 0.25 |
ENSMUST00000233304.2
ENSMUST00000233373.2 ENSMUST00000233874.2 ENSMUST00000233902.2 |
Vmn2r56
|
vomeronasal 2, receptor 56 |
| chr15_+_11000802 | 0.25 |
ENSMUST00000117100.4
|
Slc45a2
|
solute carrier family 45, member 2 |
| chr7_-_29931612 | 0.24 |
ENSMUST00000006254.6
|
Tbcb
|
tubulin folding cofactor B |
| chr9_-_101076198 | 0.22 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr7_+_19927635 | 0.22 |
ENSMUST00000168984.2
|
Vmn1r95
|
vomeronasal 1 receptor, 95 |
| chr2_-_165129773 | 0.21 |
ENSMUST00000109300.9
|
Slc35c2
|
solute carrier family 35, member C2 |
| chr7_+_106413336 | 0.21 |
ENSMUST00000166880.3
ENSMUST00000075414.8 |
Olfr701
|
olfactory receptor 701 |
| chr2_+_87362140 | 0.21 |
ENSMUST00000217113.2
|
Olfr153
|
olfactory receptor 153 |
| chr14_-_33700719 | 0.21 |
ENSMUST00000166737.2
|
Zfp488
|
zinc finger protein 488 |
| chr9_+_121196034 | 0.21 |
ENSMUST00000210636.2
ENSMUST00000045903.8 |
Trak1
|
trafficking protein, kinesin binding 1 |
| chr3_+_57332735 | 0.21 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr7_+_21006104 | 0.20 |
ENSMUST00000098739.3
|
Vmn1r125
|
vomeronasal 1 receptor 125 |
| chr8_-_96301825 | 0.20 |
ENSMUST00000180075.2
|
Prss54
|
protease, serine 54 |
| chr1_-_125362321 | 0.19 |
ENSMUST00000191544.7
|
Actr3
|
ARP3 actin-related protein 3 |
| chr5_+_31609772 | 0.18 |
ENSMUST00000202061.4
ENSMUST00000076264.8 ENSMUST00000201450.4 |
Zfp512
|
zinc finger protein 512 |
| chrX_+_110920704 | 0.18 |
ENSMUST00000131304.2
|
Tex16
|
testis expressed gene 16 |
| chr1_+_59296065 | 0.18 |
ENSMUST00000160662.8
ENSMUST00000114248.3 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr7_-_19967753 | 0.17 |
ENSMUST00000168580.2
|
Vmn1r242
|
vomeronasal 1 receptor 242 |
| chr11_-_109188917 | 0.17 |
ENSMUST00000106704.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr6_+_41098273 | 0.16 |
ENSMUST00000103270.4
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr3_+_108561247 | 0.16 |
ENSMUST00000124384.8
ENSMUST00000029483.15 |
Clcc1
|
chloride channel CLIC-like 1 |
| chr6_+_56933451 | 0.15 |
ENSMUST00000096612.4
|
Vmn1r4
|
vomeronasal 1 receptor 4 |
| chr4_+_146586445 | 0.15 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr2_+_36674288 | 0.14 |
ENSMUST00000213498.2
ENSMUST00000214909.3 ENSMUST00000215199.2 |
Olfr348
|
olfactory receptor 348 |
| chr5_+_31609802 | 0.14 |
ENSMUST00000202244.4
|
Zfp512
|
zinc finger protein 512 |
| chr19_+_13628455 | 0.13 |
ENSMUST00000213900.2
|
Olfr1490
|
olfactory receptor 1490 |
| chr17_+_18801989 | 0.13 |
ENSMUST00000165692.8
|
Vmn2r96
|
vomeronasal 2, receptor 96 |
| chr14_-_41802974 | 0.13 |
ENSMUST00000179947.2
|
Gm3543
|
predicted gene 3543 |
| chr2_-_27354006 | 0.13 |
ENSMUST00000164296.8
|
Brd3
|
bromodomain containing 3 |
| chr9_+_19620729 | 0.13 |
ENSMUST00000217450.4
ENSMUST00000212013.4 |
Olfr857
|
olfactory receptor 857 |
| chr16_-_88609108 | 0.13 |
ENSMUST00000232664.2
|
Gm20741
|
predicted gene, 20741 |
| chr2_+_86655007 | 0.13 |
ENSMUST00000217509.2
|
Olfr1094
|
olfactory receptor 1094 |
| chr19_+_13208692 | 0.13 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr6_-_122317457 | 0.13 |
ENSMUST00000160843.8
|
Phc1
|
polyhomeotic 1 |
| chrX_-_52298601 | 0.12 |
ENSMUST00000174390.2
|
Plac1
|
placental specific protein 1 |
| chr11_+_50905063 | 0.12 |
ENSMUST00000217480.2
ENSMUST00000215409.2 |
Olfr54
|
olfactory receptor 54 |
| chr7_-_37970734 | 0.12 |
ENSMUST00000032585.8
|
Pop4
|
processing of precursor 4, ribonuclease P/MRP family, (S. cerevisiae) |
| chr18_+_69633741 | 0.12 |
ENSMUST00000207214.2
ENSMUST00000201094.4 ENSMUST00000200703.4 ENSMUST00000202765.4 |
Tcf4
|
transcription factor 4 |
| chr1_-_75208734 | 0.11 |
ENSMUST00000180101.3
|
A630095N17Rik
|
RIKEN cDNA A630095N17 gene |
| chr19_-_33764859 | 0.11 |
ENSMUST00000148137.9
|
Lipo1
|
lipase, member O1 |
| chr3_-_113367891 | 0.10 |
ENSMUST00000142505.9
|
Amy1
|
amylase 1, salivary |
| chr16_-_59036978 | 0.10 |
ENSMUST00000099657.5
|
Olfr199
|
olfactory receptor 199 |
| chr1_-_97689263 | 0.10 |
ENSMUST00000171129.8
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr12_-_100125860 | 0.09 |
ENSMUST00000021596.9
ENSMUST00000221954.2 |
Nrde2
|
nrde-2 necessary for RNA interference, domain containing |
| chr5_+_26462689 | 0.09 |
ENSMUST00000074148.7
|
Gm7361
|
predicted gene 7361 |
| chr18_+_13139992 | 0.09 |
ENSMUST00000041676.3
ENSMUST00000234084.2 ENSMUST00000234565.2 |
Hrh4
|
histamine receptor H4 |
| chr19_-_13075176 | 0.09 |
ENSMUST00000208913.2
ENSMUST00000215229.2 |
Olfr1457
|
olfactory receptor 1457 |
| chr3_+_108561294 | 0.09 |
ENSMUST00000106613.2
|
Clcc1
|
chloride channel CLIC-like 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.7 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.8 | 5.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.6 | 1.9 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.5 | 1.6 | GO:0048789 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.5 | 2.6 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.5 | 4.2 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.4 | 2.6 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.4 | 2.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.3 | 0.9 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.2 | 1.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.5 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.2 | 1.8 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 | 5.0 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.2 | 2.4 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 0.8 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 1.8 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.2 | 1.4 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.9 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 0.5 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.1 | 0.8 | GO:0015819 | lysine transport(GO:0015819) |
| 0.1 | 2.1 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 3.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 2.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.3 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.1 | 0.5 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.1 | 0.6 | GO:0052805 | imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 1.2 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.5 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 3.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.3 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 1.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.2 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.3 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 6.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.8 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.2 | GO:1904171 | negative regulation of bleb assembly(GO:1904171) |
| 0.0 | 0.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) maintenance of organ identity(GO:0048496) |
| 0.0 | 0.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 2.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.8 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.0 | 1.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 2.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 3.0 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.0 | 0.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 1.4 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.8 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 2.7 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.6 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.2 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.6 | GO:0051452 | intracellular pH reduction(GO:0051452) |
| 0.0 | 0.7 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.5 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.4 | 2.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 3.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 2.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.3 | 3.0 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.2 | 0.7 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.2 | 0.8 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 6.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 4.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 2.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 3.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 4.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.9 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.3 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.2 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 5.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 6.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.5 | 3.0 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.5 | 1.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 3.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.4 | 4.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.4 | 3.0 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.4 | 2.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 1.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 2.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 6.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 1.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 5.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 0.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 0.8 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.2 | 2.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.8 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.4 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 2.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 0.1 | 1.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.4 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 2.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.4 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 0.2 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 2.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.7 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 1.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.6 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 3.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 2.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.9 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 5.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 4.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 3.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 2.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 2.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 5.0 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |