Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Hoxb3
Z-value: 1.13
Transcription factors associated with Hoxb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb3
|
ENSMUSG00000048763.12 | Hoxb3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb3 | mm39_v1_chr11_+_96214078_96214152 | -0.00 | 9.8e-01 | Click! |
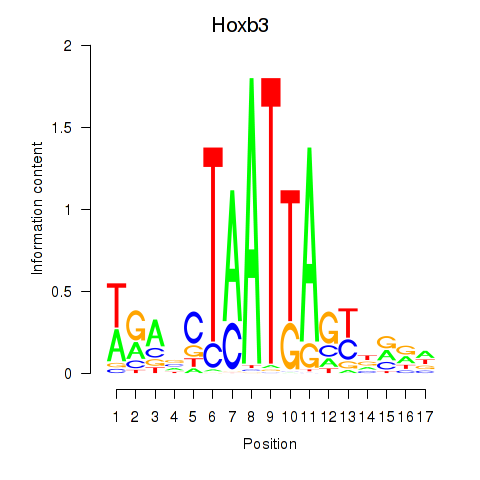
Activity profile of Hoxb3 motif
Sorted Z-values of Hoxb3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_70383976 | 12.82 |
ENSMUST00000103393.2
|
Igkv6-15
|
immunoglobulin kappa variable 6-15 |
| chr6_-_69704122 | 11.93 |
ENSMUST00000103364.3
|
Igkv5-48
|
immunoglobulin kappa variable 5-48 |
| chr6_-_69282389 | 9.75 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr12_-_113790741 | 8.71 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr6_-_69415741 | 7.83 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr5_-_134975773 | 7.36 |
ENSMUST00000051401.4
|
Cldn4
|
claudin 4 |
| chr2_-_32277773 | 6.68 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr5_-_143963413 | 6.60 |
ENSMUST00000031622.13
|
Ocm
|
oncomodulin |
| chr12_-_114672701 | 6.56 |
ENSMUST00000103505.3
ENSMUST00000193855.2 |
Ighv1-19
|
immunoglobulin heavy variable V1-19 |
| chr19_+_58748132 | 6.41 |
ENSMUST00000026081.5
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr12_-_114117264 | 6.11 |
ENSMUST00000103461.5
|
Ighv7-3
|
immunoglobulin heavy variable 7-3 |
| chr6_+_68518603 | 6.00 |
ENSMUST00000168090.3
ENSMUST00000103326.3 |
Igkv1-99
|
immunoglobulin kappa variable 1-99 |
| chr6_-_69394425 | 5.77 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chr14_+_99536111 | 5.23 |
ENSMUST00000005279.8
|
Klf5
|
Kruppel-like factor 5 |
| chr9_+_5298669 | 5.19 |
ENSMUST00000238505.2
|
Casp1
|
caspase 1 |
| chr8_-_58106057 | 4.96 |
ENSMUST00000034021.12
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chrX_+_56257374 | 4.84 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr3_-_14873406 | 4.82 |
ENSMUST00000181860.8
ENSMUST00000144327.3 |
Car1
|
carbonic anhydrase 1 |
| chr6_-_68887922 | 4.74 |
ENSMUST00000103337.3
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr8_-_110688716 | 4.71 |
ENSMUST00000001722.14
ENSMUST00000051430.7 |
Marveld3
|
MARVEL (membrane-associating) domain containing 3 |
| chr6_-_69553484 | 4.68 |
ENSMUST00000103357.4
|
Igkv4-57
|
immunoglobulin kappa variable 4-57 |
| chr2_-_32278245 | 4.63 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr9_+_53678801 | 4.51 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr13_+_19528728 | 4.36 |
ENSMUST00000179181.3
|
Trgc4
|
T cell receptor gamma, constant 4 |
| chr6_-_68994064 | 4.31 |
ENSMUST00000103341.4
|
Igkv4-80
|
immunoglobulin kappa variable 4-80 |
| chr3_-_15902583 | 4.24 |
ENSMUST00000108354.8
ENSMUST00000108349.2 ENSMUST00000108352.9 ENSMUST00000108350.8 ENSMUST00000050623.11 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr6_-_69245427 | 4.15 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr19_-_5610628 | 4.11 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr8_-_58106027 | 4.08 |
ENSMUST00000110316.3
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr5_-_143963461 | 4.02 |
ENSMUST00000110702.2
|
Ocm
|
oncomodulin |
| chr10_-_75946790 | 3.96 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr10_+_75983285 | 3.82 |
ENSMUST00000020450.4
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr5_+_122344854 | 3.74 |
ENSMUST00000145854.8
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr6_+_41928559 | 3.70 |
ENSMUST00000031898.5
|
Sval1
|
seminal vesicle antigen-like 1 |
| chr6_-_68887957 | 3.67 |
ENSMUST00000200454.2
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr2_+_118877594 | 3.64 |
ENSMUST00000152380.8
ENSMUST00000099542.9 |
Knl1
|
kinetochore scaffold 1 |
| chr4_-_119151717 | 3.64 |
ENSMUST00000079644.13
|
Ybx1
|
Y box protein 1 |
| chr4_+_3938881 | 3.58 |
ENSMUST00000108386.8
ENSMUST00000121110.8 ENSMUST00000149544.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr6_-_69678271 | 3.56 |
ENSMUST00000103363.2
|
Igkv4-50
|
immunoglobulin kappa variable 4-50 |
| chr6_-_125357756 | 3.52 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr1_+_182392577 | 3.44 |
ENSMUST00000048941.14
|
Capn8
|
calpain 8 |
| chr10_+_115854118 | 3.39 |
ENSMUST00000063470.11
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr11_-_94673526 | 3.35 |
ENSMUST00000100554.8
|
Tmem92
|
transmembrane protein 92 |
| chr6_-_69162381 | 3.27 |
ENSMUST00000103344.3
|
Igkv4-74
|
immunoglobulin kappa variable 4-74 |
| chr2_+_118877610 | 3.13 |
ENSMUST00000153300.8
ENSMUST00000028799.12 |
Knl1
|
kinetochore scaffold 1 |
| chr10_-_128361731 | 3.07 |
ENSMUST00000026427.8
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr6_-_130106861 | 2.97 |
ENSMUST00000014476.6
|
Klra8
|
killer cell lectin-like receptor, subfamily A, member 8 |
| chr4_+_3938903 | 2.76 |
ENSMUST00000121210.8
ENSMUST00000121651.8 ENSMUST00000041122.11 ENSMUST00000120732.8 ENSMUST00000119307.8 ENSMUST00000123769.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr8_-_41494890 | 2.75 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr11_-_103247150 | 2.74 |
ENSMUST00000136491.3
ENSMUST00000107023.3 |
Arhgap27
|
Rho GTPase activating protein 27 |
| chr9_-_119170456 | 2.68 |
ENSMUST00000139870.2
|
Myd88
|
myeloid differentiation primary response gene 88 |
| chr6_-_129449739 | 2.67 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr13_-_74956640 | 2.65 |
ENSMUST00000231578.2
|
Cast
|
calpastatin |
| chr1_-_192880260 | 2.61 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr13_-_74956030 | 2.47 |
ENSMUST00000065629.6
|
Cast
|
calpastatin |
| chr6_-_68784692 | 2.46 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr5_+_63969706 | 2.46 |
ENSMUST00000081747.8
ENSMUST00000196575.5 |
0610040J01Rik
|
RIKEN cDNA 0610040J01 gene |
| chr13_+_23966524 | 2.46 |
ENSMUST00000074067.4
|
Trim38
|
tripartite motif-containing 38 |
| chr3_-_15491482 | 2.41 |
ENSMUST00000099201.9
ENSMUST00000194144.3 ENSMUST00000192700.3 |
Sirpb1a
|
signal-regulatory protein beta 1A |
| chr11_+_43572825 | 2.38 |
ENSMUST00000061070.6
ENSMUST00000094294.5 |
Pwwp2a
|
PWWP domain containing 2A |
| chr19_-_32038838 | 2.37 |
ENSMUST00000096119.5
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr16_+_35861554 | 2.33 |
ENSMUST00000042203.10
|
Wdr5b
|
WD repeat domain 5B |
| chr3_-_15640045 | 2.32 |
ENSMUST00000192382.6
ENSMUST00000195778.3 ENSMUST00000091319.7 |
Sirpb1b
|
signal-regulatory protein beta 1B |
| chr15_+_10952418 | 2.32 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr1_+_157334347 | 2.31 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr4_-_3938352 | 2.30 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr1_+_157334298 | 2.24 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr10_+_128173603 | 2.23 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase |
| chr2_+_36120438 | 2.21 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr17_+_88748139 | 2.19 |
ENSMUST00000112238.9
ENSMUST00000155640.2 |
Foxn2
|
forkhead box N2 |
| chrX_+_106299484 | 2.16 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr10_+_26648473 | 2.15 |
ENSMUST00000039557.9
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr1_-_144427302 | 2.12 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr12_-_55061117 | 2.10 |
ENSMUST00000172875.8
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr5_-_137784912 | 2.06 |
ENSMUST00000031740.16
|
Mepce
|
methylphosphate capping enzyme |
| chr15_-_9529898 | 2.03 |
ENSMUST00000228782.2
ENSMUST00000003981.6 |
Il7r
|
interleukin 7 receptor |
| chr11_-_99482165 | 2.03 |
ENSMUST00000104930.2
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr1_+_40363701 | 2.00 |
ENSMUST00000095020.9
ENSMUST00000194296.6 |
Il1rl2
|
interleukin 1 receptor-like 2 |
| chr10_+_127502986 | 1.99 |
ENSMUST00000118728.8
|
Nemp1
|
nuclear envelope integral membrane protein 1 |
| chr16_+_44632096 | 1.95 |
ENSMUST00000176819.8
ENSMUST00000176321.8 |
Cd200r4
|
CD200 receptor 4 |
| chr10_+_102210290 | 1.90 |
ENSMUST00000120748.2
|
Mgat4c
|
MGAT4 family, member C |
| chr14_+_26722319 | 1.82 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr18_+_12466876 | 1.74 |
ENSMUST00000092070.13
|
Lama3
|
laminin, alpha 3 |
| chr2_+_71219561 | 1.73 |
ENSMUST00000028408.3
|
Hat1
|
histone aminotransferase 1 |
| chr16_+_22676589 | 1.73 |
ENSMUST00000004574.14
ENSMUST00000178320.2 ENSMUST00000166487.10 |
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr4_-_140501507 | 1.58 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr10_+_32959472 | 1.57 |
ENSMUST00000095762.5
ENSMUST00000218281.2 ENSMUST00000217779.2 ENSMUST00000219665.2 ENSMUST00000219931.2 |
Trdn
|
triadin |
| chr6_-_69020489 | 1.53 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chr5_-_137784943 | 1.47 |
ENSMUST00000132726.2
|
Mepce
|
methylphosphate capping enzyme |
| chr2_+_69050315 | 1.46 |
ENSMUST00000005364.12
ENSMUST00000112317.3 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr17_-_7620095 | 1.44 |
ENSMUST00000115747.3
|
Ttll2
|
tubulin tyrosine ligase-like family, member 2 |
| chr2_+_89808124 | 1.41 |
ENSMUST00000061830.2
|
Olfr1260
|
olfactory receptor 1260 |
| chr7_-_44778050 | 1.39 |
ENSMUST00000209711.2
ENSMUST00000211037.2 ENSMUST00000209927.2 ENSMUST00000209815.2 ENSMUST00000210918.2 ENSMUST00000150350.9 |
Rpl13a
|
ribosomal protein L13A |
| chr1_+_54289833 | 1.37 |
ENSMUST00000027128.11
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr1_-_171854818 | 1.36 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr5_-_137529465 | 1.35 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr19_-_40371016 | 1.33 |
ENSMUST00000225766.3
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr4_-_131802561 | 1.32 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr6_-_112364974 | 1.25 |
ENSMUST00000238755.2
ENSMUST00000060847.6 |
Ssu2
|
ssu-2 homolog (C. elegans) |
| chr7_-_44741622 | 1.23 |
ENSMUST00000210469.2
ENSMUST00000211352.2 ENSMUST00000019683.11 |
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr2_-_157179344 | 1.21 |
ENSMUST00000109536.8
|
Ghrh
|
growth hormone releasing hormone |
| chr11_+_95275458 | 1.21 |
ENSMUST00000021243.16
ENSMUST00000146556.2 |
Slc35b1
|
solute carrier family 35, member B1 |
| chr17_-_33904386 | 1.21 |
ENSMUST00000087582.13
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr5_+_140593075 | 1.18 |
ENSMUST00000031555.3
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr4_-_131802606 | 1.17 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr19_-_56378309 | 1.17 |
ENSMUST00000166203.2
ENSMUST00000167239.8 |
Nrap
|
nebulin-related anchoring protein |
| chr5_+_137015873 | 1.14 |
ENSMUST00000004968.11
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr9_-_50472620 | 1.12 |
ENSMUST00000217236.2
|
Tex12
|
testis expressed 12 |
| chr5_-_137015683 | 1.09 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr14_-_59602859 | 1.09 |
ENSMUST00000161031.2
|
Phf11d
|
PHD finger protein 11D |
| chr8_+_23901506 | 1.08 |
ENSMUST00000033952.8
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr11_-_45845992 | 1.05 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr11_+_49327451 | 1.05 |
ENSMUST00000215226.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr9_-_50472605 | 1.01 |
ENSMUST00000034568.7
|
Tex12
|
testis expressed 12 |
| chr2_+_85648823 | 1.00 |
ENSMUST00000214416.2
|
Olfr1018
|
olfactory receptor 1018 |
| chr2_+_127696548 | 0.99 |
ENSMUST00000028859.8
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr7_-_44741609 | 0.95 |
ENSMUST00000210734.2
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr6_-_23650205 | 0.95 |
ENSMUST00000115354.2
|
Rnf133
|
ring finger protein 133 |
| chr14_+_62529924 | 0.93 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr14_-_59602882 | 0.93 |
ENSMUST00000160425.8
ENSMUST00000095157.11 |
Phf11d
|
PHD finger protein 11D |
| chr14_+_54701594 | 0.93 |
ENSMUST00000022782.10
|
Lrp10
|
low-density lipoprotein receptor-related protein 10 |
| chr5_-_82272549 | 0.93 |
ENSMUST00000188072.2
ENSMUST00000185410.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr13_-_23929490 | 0.93 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3 |
| chr10_+_115979787 | 0.91 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr6_+_65019793 | 0.91 |
ENSMUST00000204696.3
|
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr7_-_44741138 | 0.89 |
ENSMUST00000210527.2
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chrX_+_13147209 | 0.87 |
ENSMUST00000000804.7
|
Ddx3x
|
DEAD box helicase 3, X-linked |
| chr6_-_115014777 | 0.86 |
ENSMUST00000174848.8
ENSMUST00000032461.12 |
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr17_-_33904345 | 0.85 |
ENSMUST00000234474.2
ENSMUST00000139302.8 ENSMUST00000114385.9 |
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr11_-_102076028 | 0.85 |
ENSMUST00000107156.9
ENSMUST00000021297.6 |
Lsm12
|
LSM12 homolog |
| chr1_-_65102460 | 0.84 |
ENSMUST00000146122.2
ENSMUST00000045028.15 |
Crygd
|
crystallin, gamma D |
| chr4_-_14621669 | 0.84 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr9_+_119170486 | 0.83 |
ENSMUST00000175743.8
ENSMUST00000176397.8 |
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr2_+_110551976 | 0.83 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
| chr5_-_138169253 | 0.78 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr9_+_40712562 | 0.75 |
ENSMUST00000117557.8
|
Hspa8
|
heat shock protein 8 |
| chr6_-_41752111 | 0.74 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr11_+_116734104 | 0.74 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr11_+_58468556 | 0.72 |
ENSMUST00000203418.4
|
Olfr325
|
olfactory receptor 325 |
| chrX_+_101847829 | 0.71 |
ENSMUST00000131451.2
|
Dmrtc1c1
|
DMRT-like family C1c1 |
| chrX_+_162694397 | 0.70 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr11_-_99121822 | 0.69 |
ENSMUST00000103133.4
|
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr13_-_57043550 | 0.67 |
ENSMUST00000022023.13
ENSMUST00000174457.9 ENSMUST00000109871.8 |
Trpc7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr5_+_33493529 | 0.67 |
ENSMUST00000202113.2
|
Maea
|
macrophage erythroblast attacher |
| chr10_+_127919142 | 0.66 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr7_-_24371457 | 0.65 |
ENSMUST00000078001.7
|
Tex101
|
testis expressed gene 101 |
| chr13_-_58506890 | 0.64 |
ENSMUST00000225388.2
|
Kif27
|
kinesin family member 27 |
| chr4_-_41045381 | 0.64 |
ENSMUST00000054945.8
|
Aqp7
|
aquaporin 7 |
| chr3_-_75072319 | 0.63 |
ENSMUST00000124618.2
|
Zbbx
|
zinc finger, B-box domain containing |
| chr14_+_26414422 | 0.63 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr12_-_99529767 | 0.61 |
ENSMUST00000176928.3
ENSMUST00000223484.2 |
Foxn3
|
forkhead box N3 |
| chr2_-_111843053 | 0.61 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr18_+_36868084 | 0.60 |
ENSMUST00000007046.9
ENSMUST00000237870.2 |
Tmco6
|
transmembrane and coiled-coil domains 6 |
| chr10_-_128885867 | 0.60 |
ENSMUST00000216460.2
|
Olfr765
|
olfactory receptor 765 |
| chr2_+_110551685 | 0.58 |
ENSMUST00000111016.9
|
Muc15
|
mucin 15 |
| chr15_-_38079089 | 0.56 |
ENSMUST00000110336.4
|
Ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr12_+_112727089 | 0.56 |
ENSMUST00000063888.5
|
Pld4
|
phospholipase D family, member 4 |
| chr9_+_21634779 | 0.56 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr7_+_29683373 | 0.55 |
ENSMUST00000148442.8
|
Zfp568
|
zinc finger protein 568 |
| chr6_-_129752812 | 0.55 |
ENSMUST00000095409.3
|
Klrh1
|
killer cell lectin-like receptor subfamily H, member 1 |
| chr9_+_20209828 | 0.55 |
ENSMUST00000215540.2
ENSMUST00000075717.7 |
Olfr873
|
olfactory receptor 873 |
| chr9_+_39933648 | 0.55 |
ENSMUST00000059859.5
|
Olfr981
|
olfactory receptor 981 |
| chr11_+_58668915 | 0.54 |
ENSMUST00000081533.5
|
Olfr315
|
olfactory receptor 315 |
| chr11_+_69471185 | 0.53 |
ENSMUST00000171247.8
ENSMUST00000108658.10 ENSMUST00000005371.12 |
Trp53
|
transformation related protein 53 |
| chr6_-_68968278 | 0.51 |
ENSMUST00000197966.2
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chr2_+_110551927 | 0.50 |
ENSMUST00000111017.9
|
Muc15
|
mucin 15 |
| chr4_-_43710231 | 0.50 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr18_+_52958382 | 0.50 |
ENSMUST00000238707.2
|
Gm50457
|
predicted gene, 50457 |
| chr2_-_87868043 | 0.47 |
ENSMUST00000129056.3
|
Olfr73
|
olfactory receptor 73 |
| chr6_-_23650297 | 0.46 |
ENSMUST00000063548.4
|
Rnf133
|
ring finger protein 133 |
| chr13_-_112698563 | 0.45 |
ENSMUST00000224510.2
|
Il31ra
|
interleukin 31 receptor A |
| chr11_+_5738480 | 0.45 |
ENSMUST00000109845.8
ENSMUST00000020769.14 ENSMUST00000102928.5 |
Dbnl
|
drebrin-like |
| chr7_-_102143980 | 0.44 |
ENSMUST00000058750.4
|
Olfr545
|
olfactory receptor 545 |
| chr17_-_37591309 | 0.42 |
ENSMUST00000077585.3
|
Olfr99
|
olfactory receptor 99 |
| chr11_+_101556367 | 0.41 |
ENSMUST00000039388.3
|
Arl4d
|
ADP-ribosylation factor-like 4D |
| chrX_+_113814692 | 0.41 |
ENSMUST00000059509.3
|
Ube2dnl1
|
ubiquitin-conjugating enzyme E2D N-terminal like 1 |
| chr7_+_45271229 | 0.41 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr6_+_40419797 | 0.41 |
ENSMUST00000038907.9
ENSMUST00000141490.2 |
Wee2
|
WEE1 homolog 2 (S. pombe) |
| chr12_-_111150925 | 0.39 |
ENSMUST00000121608.2
|
4930595D18Rik
|
RIKEN cDNA 4930595D18 gene |
| chr14_-_52704952 | 0.38 |
ENSMUST00000206520.3
|
Olfr1508
|
olfactory receptor 1508 |
| chr2_-_85193402 | 0.38 |
ENSMUST00000111597.3
|
Olfr988
|
olfactory receptor 988 |
| chr11_-_95966407 | 0.37 |
ENSMUST00000107686.8
ENSMUST00000107684.2 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr2_-_111820618 | 0.36 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr6_+_65019574 | 0.35 |
ENSMUST00000031984.9
ENSMUST00000205118.3 |
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr2_+_32178325 | 0.35 |
ENSMUST00000100194.10
ENSMUST00000131712.9 ENSMUST00000081670.14 ENSMUST00000147707.8 ENSMUST00000129193.3 |
Golga2
|
golgi autoantigen, golgin subfamily a, 2 |
| chr13_-_21900313 | 0.35 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr16_-_59166089 | 0.34 |
ENSMUST00000084791.4
|
Olfr206
|
olfactory receptor 206 |
| chr2_+_68490186 | 0.34 |
ENSMUST00000055930.6
|
4932414N04Rik
|
RIKEN cDNA 4932414N04 gene |
| chr5_-_137213788 | 0.34 |
ENSMUST00000239135.2
|
Gm31160
|
predicted gene, 31160 |
| chrX_-_138683102 | 0.32 |
ENSMUST00000101217.4
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr14_-_50425655 | 0.32 |
ENSMUST00000205837.3
|
Olfr730
|
olfactory receptor 730 |
| chr9_+_40092216 | 0.32 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr12_-_83534482 | 0.31 |
ENSMUST00000177959.8
ENSMUST00000178756.8 |
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chrY_+_5656986 | 0.30 |
ENSMUST00000190391.2
|
Gm21854
|
predicted gene, 21854 |
| chr14_+_62901114 | 0.30 |
ENSMUST00000171692.2
|
Serpine3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr2_+_129965195 | 0.30 |
ENSMUST00000028888.5
|
Tgm6
|
transglutaminase 6 |
| chr19_+_12364643 | 0.30 |
ENSMUST00000217062.3
ENSMUST00000216145.2 ENSMUST00000213657.2 |
Olfr1440
|
olfactory receptor 1440 |
| chr14_-_75991903 | 0.29 |
ENSMUST00000049168.9
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr9_+_18944031 | 0.28 |
ENSMUST00000213018.3
|
Olfr835
|
olfactory receptor 835 |
| chr2_-_90301592 | 0.28 |
ENSMUST00000111493.8
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 2.1 | 10.6 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 1.6 | 4.8 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 1.4 | 4.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 1.3 | 5.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.7 | 3.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.7 | 4.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.7 | 2.7 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.6 | 7.8 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.5 | 1.9 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.5 | 4.5 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.4 | 6.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.4 | 1.2 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.4 | 5.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.4 | 3.5 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.4 | 6.4 | GO:0019374 | galactolipid metabolic process(GO:0019374) |
| 0.4 | 1.1 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.4 | 2.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.3 | 2.0 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.3 | 1.6 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 2.2 | GO:0035633 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.3 | 2.3 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.3 | 74.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.3 | 5.1 | GO:0007343 | egg activation(GO:0007343) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.3 | 1.6 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.3 | 7.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 3.7 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.2 | 0.7 | GO:0097212 | protein targeting to vacuole involved in autophagy(GO:0071211) lysosomal membrane organization(GO:0097212) positive regulation of protein folding(GO:1903334) |
| 0.2 | 4.7 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.2 | 1.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.2 | 1.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 2.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.2 | 0.7 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.2 | 0.4 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.2 | 0.6 | GO:1903978 | regulation of microglial cell activation(GO:1903978) |
| 0.2 | 0.5 | GO:1902689 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 0.2 | 1.8 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 1.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 9.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 0.6 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.1 | 2.5 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.7 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.6 | GO:0015793 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.1 | 0.9 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 15.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 2.3 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 1.1 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.1 | 4.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 2.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.9 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 3.0 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.9 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 0.4 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.1 | 1.5 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 1.3 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 2.2 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 1.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.3 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.6 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 2.3 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 0.4 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 2.0 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 3.4 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.8 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.8 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 1.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 1.6 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 2.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0043415 | vitamin A metabolic process(GO:0006776) positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 1.1 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 2.1 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.6 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 4.0 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.6 | 3.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.5 | 2.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.5 | 1.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.4 | 1.7 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.4 | 1.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.3 | 6.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.3 | 1.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 1.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 7.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 2.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 0.6 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.2 | 2.1 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 6.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.2 | 0.9 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 1.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 3.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.7 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 15.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.7 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 98.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.5 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 1.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 1.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 2.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.7 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 2.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 24.0 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 4.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.7 | 5.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.7 | 2.7 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.6 | 7.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.6 | 3.7 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.5 | 2.0 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.5 | 4.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.4 | 5.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 9.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 6.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 1.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 1.5 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.4 | 1.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.3 | 2.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.3 | 1.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 1.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.3 | 2.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 0.9 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.3 | 0.8 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.2 | 1.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.2 | 1.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.2 | 0.7 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.2 | 1.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 0.7 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 33.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 4.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.0 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.9 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 2.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.2 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 1.5 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 2.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.7 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 11.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 3.5 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.4 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 3.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 2.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.0 | 2.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 4.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 3.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 3.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 4.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 6.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 2.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 4.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 3.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 7.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 2.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 10.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 5.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 7.0 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.1 | 2.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 4.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 2.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 2.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 4.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 5.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |