Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Hoxb4
Z-value: 0.54
Transcription factors associated with Hoxb4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb4
|
ENSMUSG00000038692.9 | Hoxb4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb4 | mm39_v1_chr11_+_96209093_96209093 | 0.02 | 8.6e-01 | Click! |
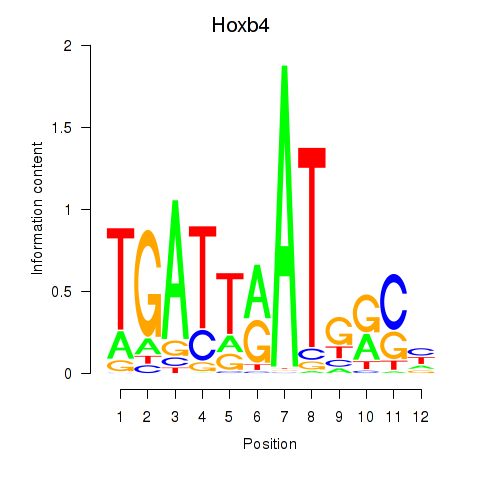
Activity profile of Hoxb4 motif
Sorted Z-values of Hoxb4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_20329823 | 3.70 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr7_+_130633776 | 3.06 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr16_+_78727829 | 2.76 |
ENSMUST00000114216.2
ENSMUST00000069148.13 ENSMUST00000023568.14 |
Chodl
|
chondrolectin |
| chr8_-_62576140 | 2.65 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr2_-_84605764 | 2.64 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr10_-_88440869 | 2.48 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr2_-_84605732 | 2.47 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr10_-_88440996 | 1.98 |
ENSMUST00000121629.8
|
Mybpc1
|
myosin binding protein C, slow-type |
| chr1_+_106866678 | 1.71 |
ENSMUST00000112724.3
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr6_+_116627567 | 1.47 |
ENSMUST00000067354.10
ENSMUST00000178241.4 |
Depp1
|
DEPP1 autophagy regulator |
| chr4_+_58285957 | 1.04 |
ENSMUST00000081919.12
ENSMUST00000177951.8 ENSMUST00000098059.10 ENSMUST00000179951.2 ENSMUST00000102893.10 ENSMUST00000084578.12 ENSMUST00000098057.10 |
Musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr6_+_116627635 | 0.91 |
ENSMUST00000204555.2
|
Depp1
|
DEPP1 autophagy regulator |
| chr3_-_144804784 | 0.84 |
ENSMUST00000040465.11
ENSMUST00000198993.2 |
Clca2
|
chloride channel accessory 2 |
| chr18_-_56695288 | 0.83 |
ENSMUST00000170309.8
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr11_-_40646090 | 0.83 |
ENSMUST00000020576.8
|
Ccng1
|
cyclin G1 |
| chr13_+_94219934 | 0.83 |
ENSMUST00000156071.2
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr8_+_26298502 | 0.80 |
ENSMUST00000033979.6
|
Star
|
steroidogenic acute regulatory protein |
| chr1_-_171050077 | 0.79 |
ENSMUST00000005817.9
|
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr18_-_56695333 | 0.79 |
ENSMUST00000066208.13
ENSMUST00000172734.8 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr2_-_38604503 | 0.77 |
ENSMUST00000028084.5
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr18_-_56695259 | 0.75 |
ENSMUST00000171844.3
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr1_-_171050004 | 0.69 |
ENSMUST00000147246.2
ENSMUST00000111326.8 ENSMUST00000138184.8 |
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr1_-_37996838 | 0.66 |
ENSMUST00000027254.10
ENSMUST00000114894.2 |
Lyg1
|
lysozyme G-like 1 |
| chr13_-_21823691 | 0.65 |
ENSMUST00000043081.3
|
Olfr11
|
olfactory receptor 11 |
| chr16_-_19226828 | 0.65 |
ENSMUST00000052516.5
ENSMUST00000206410.3 |
Olfr165
|
olfactory receptor 165 |
| chrX_-_8059597 | 0.65 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr14_+_32972324 | 0.63 |
ENSMUST00000131086.3
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr8_+_120955195 | 0.57 |
ENSMUST00000180448.3
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr4_+_43493344 | 0.56 |
ENSMUST00000030181.12
ENSMUST00000107922.3 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr4_+_151081538 | 0.55 |
ENSMUST00000030803.2
|
Uts2
|
urotensin 2 |
| chr16_-_94171340 | 0.50 |
ENSMUST00000138514.2
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr1_-_135934080 | 0.49 |
ENSMUST00000166193.9
|
Igfn1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr10_-_59452489 | 0.44 |
ENSMUST00000020312.13
|
Mcu
|
mitochondrial calcium uniporter |
| chr8_-_106198112 | 0.43 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr10_-_89457115 | 0.42 |
ENSMUST00000020102.14
|
Slc17a8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
| chr7_+_144391786 | 0.33 |
ENSMUST00000155320.8
|
Fgf3
|
fibroblast growth factor 3 |
| chr6_+_142359099 | 0.30 |
ENSMUST00000126521.9
ENSMUST00000211094.2 |
Spx
|
spexin hormone |
| chr11_+_46701619 | 0.28 |
ENSMUST00000068877.7
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr1_+_9615619 | 0.27 |
ENSMUST00000072079.9
|
Rrs1
|
ribosome biogenesis regulator 1 |
| chr7_+_29931309 | 0.27 |
ENSMUST00000019882.16
ENSMUST00000149654.8 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr17_+_44263890 | 0.24 |
ENSMUST00000177857.9
ENSMUST00000044792.6 |
Rcan2
|
regulator of calcineurin 2 |
| chr11_+_58471639 | 0.22 |
ENSMUST00000169428.5
|
Olfr325
|
olfactory receptor 325 |
| chr7_+_107702486 | 0.21 |
ENSMUST00000104917.4
|
Olfr483
|
olfactory receptor 483 |
| chr7_+_29931735 | 0.17 |
ENSMUST00000108193.2
ENSMUST00000108192.2 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr10_-_41585293 | 0.17 |
ENSMUST00000019955.16
ENSMUST00000099932.10 |
Ccdc162
|
coiled-coil domain containing 162 |
| chr18_+_56695515 | 0.16 |
ENSMUST00000130163.8
ENSMUST00000132628.8 |
Phax
|
phosphorylated adaptor for RNA export |
| chr10_+_129376408 | 0.16 |
ENSMUST00000076575.4
|
Olfr792
|
olfactory receptor 792 |
| chr2_+_102488985 | 0.15 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr1_+_11063678 | 0.14 |
ENSMUST00000027056.12
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr2_-_118987305 | 0.13 |
ENSMUST00000135419.8
ENSMUST00000129351.2 ENSMUST00000139519.2 ENSMUST00000094695.12 |
Rmdn3
|
regulator of microtubule dynamics 3 |
| chr9_+_40098375 | 0.12 |
ENSMUST00000062229.6
|
Olfr986
|
olfactory receptor 986 |
| chr3_-_51184730 | 0.11 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr7_-_29931612 | 0.11 |
ENSMUST00000006254.6
|
Tbcb
|
tubulin folding cofactor B |
| chr12_-_54842488 | 0.11 |
ENSMUST00000005798.9
|
Snx6
|
sorting nexin 6 |
| chrX_-_142716200 | 0.10 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr3_-_97318495 | 0.10 |
ENSMUST00000060912.4
|
Olfr1402
|
olfactory receptor 1402 |
| chr4_-_35845204 | 0.10 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr7_-_103214484 | 0.08 |
ENSMUST00000106886.2
|
Olfr616
|
olfactory receptor 616 |
| chr15_+_101013704 | 0.03 |
ENSMUST00000229954.2
|
Ankrd33
|
ankyrin repeat domain 33 |
| chr7_+_16821858 | 0.03 |
ENSMUST00000152671.2
|
Psg16
|
pregnancy specific glycoprotein 16 |
| chr14_+_58308004 | 0.03 |
ENSMUST00000165526.9
|
Fgf9
|
fibroblast growth factor 9 |
| chrX_-_142716085 | 0.02 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr15_+_82230155 | 0.02 |
ENSMUST00000023086.15
|
Smdt1
|
single-pass membrane protein with aspartate rich tail 1 |
| chr4_-_118687635 | 0.01 |
ENSMUST00000076019.4
|
Olfr1333
|
olfactory receptor 1333 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.3 | 4.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 1.0 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.2 | 0.6 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.2 | 0.8 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 3.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.8 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.1 | 1.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.7 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 0.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.3 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 0.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.4 | GO:0015813 | L-glutamate transport(GO:0015813) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 4.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 5.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.6 | 2.4 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.2 | 4.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 5.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.4 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.1 | 1.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 3.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 2.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.8 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.4 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 1.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 6.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 4.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |