Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Hoxb5
Z-value: 0.79
Transcription factors associated with Hoxb5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb5
|
ENSMUSG00000038700.5 | Hoxb5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb5 | mm39_v1_chr11_+_96194333_96194378 | -0.41 | 2.9e-04 | Click! |
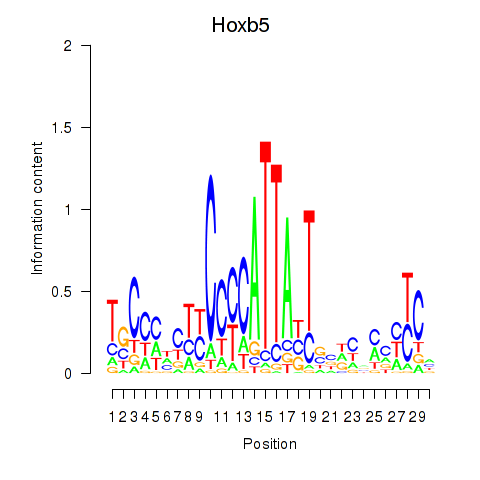
Activity profile of Hoxb5 motif
Sorted Z-values of Hoxb5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_125609440 | 8.02 |
ENSMUST00000031446.7
|
Tmem132b
|
transmembrane protein 132B |
| chr2_-_151474391 | 7.24 |
ENSMUST00000137936.2
ENSMUST00000146172.8 ENSMUST00000094456.10 ENSMUST00000148755.8 ENSMUST00000109875.8 ENSMUST00000028951.14 ENSMUST00000109877.10 |
Snph
|
syntaphilin |
| chr1_-_75240551 | 7.04 |
ENSMUST00000186178.7
ENSMUST00000189769.7 ENSMUST00000027404.12 |
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr9_-_35469818 | 6.35 |
ENSMUST00000034612.7
|
Ddx25
|
DEAD box helicase 25 |
| chr17_-_57394718 | 6.30 |
ENSMUST00000071135.6
|
Tubb4a
|
tubulin, beta 4A class IVA |
| chr7_-_44861235 | 5.81 |
ENSMUST00000210741.2
ENSMUST00000209466.2 |
Dkkl1
|
dickkopf-like 1 |
| chr19_-_45804446 | 5.68 |
ENSMUST00000079431.10
ENSMUST00000026247.13 ENSMUST00000162528.9 |
Kcnip2
|
Kv channel-interacting protein 2 |
| chr2_+_102489558 | 5.24 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr5_+_125608838 | 4.85 |
ENSMUST00000185104.2
|
Tmem132b
|
transmembrane protein 132B |
| chr7_+_3439144 | 4.73 |
ENSMUST00000182222.8
|
Cacng8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr10_-_80850712 | 4.66 |
ENSMUST00000126317.2
ENSMUST00000092285.10 ENSMUST00000117805.8 |
Gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr15_-_95553841 | 4.62 |
ENSMUST00000054244.7
|
Dbx2
|
developing brain homeobox 2 |
| chr12_+_88689638 | 4.59 |
ENSMUST00000190626.7
ENSMUST00000167103.8 |
Nrxn3
|
neurexin III |
| chr5_-_103359117 | 4.42 |
ENSMUST00000112846.8
ENSMUST00000170792.9 ENSMUST00000112847.9 ENSMUST00000238446.3 ENSMUST00000133069.8 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr16_-_44153498 | 4.15 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr7_-_108682491 | 4.00 |
ENSMUST00000120876.2
ENSMUST00000055993.13 |
Ric3
|
RIC3 acetylcholine receptor chaperone |
| chr13_+_88969591 | 3.77 |
ENSMUST00000118731.8
ENSMUST00000081769.13 |
Edil3
|
EGF-like repeats and discoidin I-like domains 3 |
| chr13_+_88969725 | 3.59 |
ENSMUST00000043111.7
|
Edil3
|
EGF-like repeats and discoidin I-like domains 3 |
| chr6_-_113478779 | 3.51 |
ENSMUST00000101059.4
ENSMUST00000204268.3 ENSMUST00000205170.2 ENSMUST00000205075.2 ENSMUST00000204134.3 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr7_-_123099672 | 3.45 |
ENSMUST00000042470.14
ENSMUST00000128217.2 |
Zkscan2
|
zinc finger with KRAB and SCAN domains 2 |
| chr7_+_16609227 | 3.43 |
ENSMUST00000108493.3
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr6_-_18514801 | 3.28 |
ENSMUST00000090601.12
|
Cttnbp2
|
cortactin binding protein 2 |
| chr9_-_52590686 | 3.26 |
ENSMUST00000098768.3
ENSMUST00000213843.2 |
AI593442
|
expressed sequence AI593442 |
| chr1_-_193052533 | 3.19 |
ENSMUST00000169907.8
|
Camk1g
|
calcium/calmodulin-dependent protein kinase I gamma |
| chr10_+_80136830 | 3.11 |
ENSMUST00000140828.8
ENSMUST00000138909.8 |
Apc2
|
APC regulator of WNT signaling pathway 2 |
| chr1_-_193052568 | 2.98 |
ENSMUST00000016323.11
|
Camk1g
|
calcium/calmodulin-dependent protein kinase I gamma |
| chrX_-_164110372 | 2.88 |
ENSMUST00000058787.9
|
Glra2
|
glycine receptor, alpha 2 subunit |
| chr18_-_16942289 | 2.87 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr12_-_79027531 | 2.62 |
ENSMUST00000174072.8
|
Tmem229b
|
transmembrane protein 229B |
| chr16_-_44153288 | 2.56 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr12_-_36092475 | 2.52 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chr1_+_74583504 | 2.44 |
ENSMUST00000027362.14
|
Plcd4
|
phospholipase C, delta 4 |
| chr5_+_143869875 | 2.34 |
ENSMUST00000166847.8
|
Rsph10b
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr8_-_71303383 | 2.32 |
ENSMUST00000212084.2
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr6_+_121613177 | 2.29 |
ENSMUST00000032203.9
|
A2m
|
alpha-2-macroglobulin |
| chrX_+_149829131 | 2.22 |
ENSMUST00000112685.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr10_-_127047396 | 2.20 |
ENSMUST00000013970.9
|
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr17_-_23990512 | 2.15 |
ENSMUST00000226460.2
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr16_+_16888145 | 2.06 |
ENSMUST00000232574.2
|
Ypel1
|
yippee like 1 |
| chr17_-_23990479 | 2.01 |
ENSMUST00000086325.13
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr1_+_132131590 | 1.97 |
ENSMUST00000185223.2
|
Lemd1
|
LEM domain containing 1 |
| chr16_+_16888084 | 1.91 |
ENSMUST00000231514.2
|
Ypel1
|
yippee like 1 |
| chrX_+_20554193 | 1.84 |
ENSMUST00000115364.8
|
Cdk16
|
cyclin-dependent kinase 16 |
| chr17_-_80885197 | 1.83 |
ENSMUST00000234602.2
|
Cdkl4
|
cyclin-dependent kinase-like 4 |
| chr8_+_95807814 | 1.79 |
ENSMUST00000034239.9
|
Katnb1
|
katanin p80 (WD40-containing) subunit B 1 |
| chrX_-_133012600 | 1.76 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr11_-_69088635 | 1.62 |
ENSMUST00000094078.4
ENSMUST00000021262.10 |
Alox8
|
arachidonate 8-lipoxygenase |
| chr2_+_158509039 | 1.61 |
ENSMUST00000045503.11
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chrX_+_106860083 | 1.54 |
ENSMUST00000143975.8
ENSMUST00000144695.8 ENSMUST00000167154.2 |
Tent5d
|
terminal nucleotidyltransferase 5D |
| chr18_-_61840654 | 1.53 |
ENSMUST00000025472.7
|
Pcyox1l
|
prenylcysteine oxidase 1 like |
| chr2_-_153067297 | 1.51 |
ENSMUST00000099194.4
|
Tspyl3
|
TSPY-like 3 |
| chrX_-_71699740 | 1.51 |
ENSMUST00000055966.13
|
Gabra3
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 |
| chr8_-_123768984 | 1.47 |
ENSMUST00000212937.2
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr9_+_8900460 | 1.44 |
ENSMUST00000070463.10
ENSMUST00000098986.4 |
Pgr
|
progesterone receptor |
| chr7_-_126807581 | 1.43 |
ENSMUST00000120705.3
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr5_+_112597272 | 1.42 |
ENSMUST00000100882.3
|
Gm6588
|
predicted gene 6588 |
| chr17_-_48002328 | 1.41 |
ENSMUST00000152214.4
ENSMUST00000113299.6 |
Prickle4
|
prickle planar cell polarity protein 4 |
| chr8_-_25086976 | 1.38 |
ENSMUST00000033956.7
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr16_+_16887991 | 1.27 |
ENSMUST00000232258.2
|
Ypel1
|
yippee like 1 |
| chr8_+_21382681 | 1.24 |
ENSMUST00000098908.4
|
Defb33
|
defensin beta 33 |
| chr2_-_164585102 | 1.21 |
ENSMUST00000103096.10
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr7_-_86044743 | 1.16 |
ENSMUST00000053958.6
|
Olfr303
|
olfactory receptor 303 |
| chr12_-_79030250 | 1.16 |
ENSMUST00000070174.14
|
Tmem229b
|
transmembrane protein 229B |
| chr8_-_26087475 | 1.16 |
ENSMUST00000210810.2
ENSMUST00000210616.2 ENSMUST00000079160.8 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr7_+_86175967 | 1.13 |
ENSMUST00000071112.4
|
Olfr297
|
olfactory receptor 297 |
| chrX_+_7688528 | 1.03 |
ENSMUST00000009875.5
|
Kcnd1
|
potassium voltage-gated channel, Shal-related family, member 1 |
| chr7_-_133310687 | 1.01 |
ENSMUST00000106146.8
|
Uros
|
uroporphyrinogen III synthase |
| chr2_-_115896279 | 0.95 |
ENSMUST00000110907.8
ENSMUST00000110908.9 |
Meis2
|
Meis homeobox 2 |
| chr9_+_107277242 | 0.92 |
ENSMUST00000166799.8
ENSMUST00000170737.3 |
Cacna2d2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chr7_-_133310779 | 0.92 |
ENSMUST00000124759.2
ENSMUST00000106144.8 ENSMUST00000106145.10 |
Uros
|
uroporphyrinogen III synthase |
| chr7_-_30850429 | 0.91 |
ENSMUST00000085636.13
ENSMUST00000001280.14 |
Gramd1a
|
GRAM domain containing 1A |
| chr4_-_149222057 | 0.89 |
ENSMUST00000030813.10
|
Cenps
|
centromere protein S |
| chr19_-_7460550 | 0.87 |
ENSMUST00000088169.7
|
Rtn3
|
reticulon 3 |
| chr8_-_123768759 | 0.84 |
ENSMUST00000098334.13
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr14_-_40726472 | 0.83 |
ENSMUST00000153830.8
|
Prxl2a
|
peroxiredoxin like 2A |
| chr8_-_26087552 | 0.79 |
ENSMUST00000210234.2
ENSMUST00000211422.2 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr2_+_125089110 | 0.79 |
ENSMUST00000082122.14
|
Dut
|
deoxyuridine triphosphatase |
| chr16_-_64422716 | 0.77 |
ENSMUST00000209382.3
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr2_+_152804405 | 0.75 |
ENSMUST00000099197.9
ENSMUST00000103155.10 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr11_-_95966477 | 0.70 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr9_+_107277058 | 0.69 |
ENSMUST00000085092.12
ENSMUST00000164988.9 |
Cacna2d2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chr4_-_149221998 | 0.67 |
ENSMUST00000176124.8
ENSMUST00000177408.2 ENSMUST00000105695.2 |
Cenps
|
centromere protein S |
| chr8_-_86567506 | 0.64 |
ENSMUST00000034140.9
|
Itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr19_-_7460600 | 0.64 |
ENSMUST00000235593.2
ENSMUST00000088171.12 ENSMUST00000065304.13 ENSMUST00000025667.14 |
Rtn3
|
reticulon 3 |
| chrX_-_103244703 | 0.62 |
ENSMUST00000087879.11
|
Nexmif
|
neurite extension and migration factor |
| chr13_-_31158031 | 0.61 |
ENSMUST00000021785.8
|
Exoc2
|
exocyst complex component 2 |
| chr9_+_20492593 | 0.54 |
ENSMUST00000115557.10
|
Zfp846
|
zinc finger protein 846 |
| chr9_-_19163273 | 0.53 |
ENSMUST00000214019.2
ENSMUST00000214267.2 |
Olfr843
|
olfactory receptor 843 |
| chr8_+_124059414 | 0.45 |
ENSMUST00000010298.7
|
Spire2
|
spire type actin nucleation factor 2 |
| chr7_+_127441119 | 0.43 |
ENSMUST00000121705.8
|
Stx4a
|
syntaxin 4A (placental) |
| chr1_+_92878566 | 0.41 |
ENSMUST00000185421.2
|
Gpr35
|
G protein-coupled receptor 35 |
| chr7_+_127440924 | 0.37 |
ENSMUST00000033075.14
|
Stx4a
|
syntaxin 4A (placental) |
| chr8_-_129525175 | 0.36 |
ENSMUST00000195093.2
|
Gm3952
|
predicted gene 3952 |
| chr2_-_166904625 | 0.35 |
ENSMUST00000128676.2
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr2_+_26863494 | 0.35 |
ENSMUST00000102891.4
|
Adamts13
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 13 |
| chr2_-_5867734 | 0.34 |
ENSMUST00000071016.3
|
Gm13199
|
predicted gene 13199 |
| chr1_-_166237341 | 0.32 |
ENSMUST00000135673.8
ENSMUST00000169324.8 ENSMUST00000128861.3 |
Pogk
|
pogo transposable element with KRAB domain |
| chr13_+_23879775 | 0.31 |
ENSMUST00000041052.5
|
H1f6
|
H1.6 linker histone, cluster member |
| chr7_-_45116316 | 0.29 |
ENSMUST00000033093.10
|
Bax
|
BCL2-associated X protein |
| chr13_-_14237958 | 0.28 |
ENSMUST00000223174.2
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr6_+_48906825 | 0.26 |
ENSMUST00000031837.8
|
Doxl1
|
diamine oxidase-like protein 1 |
| chr14_-_64225223 | 0.24 |
ENSMUST00000022532.6
|
4930578I06Rik
|
RIKEN cDNA 4930578I06 gene |
| chr2_+_26863428 | 0.23 |
ENSMUST00000014996.14
|
Adamts13
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 13 |
| chr15_+_44482545 | 0.22 |
ENSMUST00000227691.2
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chrX_-_103244784 | 0.18 |
ENSMUST00000118314.8
|
Nexmif
|
neurite extension and migration factor |
| chr10_-_61979685 | 0.18 |
ENSMUST00000141724.8
ENSMUST00000150057.2 |
Fam241b
|
family with sequence similarity 241, member B |
| chr1_-_74583434 | 0.15 |
ENSMUST00000189257.7
|
Usp37
|
ubiquitin specific peptidase 37 |
| chr7_-_103710652 | 0.14 |
ENSMUST00000074064.5
|
4930516K23Rik
|
RIKEN cDNA 4930516K23 gene |
| chr16_+_32315090 | 0.12 |
ENSMUST00000231510.2
|
Zdhhc19
|
zinc finger, DHHC domain containing 19 |
| chr6_-_128339775 | 0.12 |
ENSMUST00000112152.8
ENSMUST00000057421.15 ENSMUST00000112151.2 |
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chr5_+_117271632 | 0.12 |
ENSMUST00000179276.8
ENSMUST00000092889.12 ENSMUST00000145640.8 |
Taok3
|
TAO kinase 3 |
| chr6_+_86415342 | 0.12 |
ENSMUST00000050497.14
ENSMUST00000203568.3 |
C87436
|
expressed sequence C87436 |
| chr14_+_31881822 | 0.11 |
ENSMUST00000163336.8
ENSMUST00000169722.8 ENSMUST00000168385.8 |
Ncoa4
|
nuclear receptor coactivator 4 |
| chr7_-_45116216 | 0.10 |
ENSMUST00000210392.2
ENSMUST00000211365.2 |
Bax
|
BCL2-associated X protein |
| chr13_-_14237980 | 0.05 |
ENSMUST00000222687.2
ENSMUST00000221338.2 ENSMUST00000221713.2 ENSMUST00000170957.3 |
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr5_-_108956716 | 0.05 |
ENSMUST00000233055.2
ENSMUST00000232952.2 ENSMUST00000233745.2 ENSMUST00000172140.3 |
Vmn2r8
|
vomeronasal 2, receptor 8 |
| chr7_-_140480314 | 0.04 |
ENSMUST00000026561.10
|
Cox8b
|
cytochrome c oxidase subunit 8B |
| chr3_-_89067462 | 0.02 |
ENSMUST00000029686.4
|
Hcn3
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.7 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.8 | 2.3 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.7 | 5.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.6 | 1.9 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.6 | 4.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.6 | 6.7 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.5 | 1.5 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.5 | 4.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.5 | 1.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.4 | 1.8 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.4 | 7.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.4 | 2.9 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.4 | 1.6 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.4 | 1.6 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.4 | 1.4 | GO:1904708 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 0.3 | 1.6 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.3 | 6.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.3 | 2.9 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.3 | 0.8 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.3 | 5.8 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 | 0.8 | GO:0006226 | dUMP biosynthetic process(GO:0006226) pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.2 | 2.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 4.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 1.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 1.6 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.4 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 0.1 | 4.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 6.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.5 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.6 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.1 | 1.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 4.0 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 7.2 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.1 | 5.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 2.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.8 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 1.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 7.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 6.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 2.2 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 3.4 | GO:0006813 | potassium ion transport(GO:0006813) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 1.6 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.3 | 6.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.3 | 6.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 1.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 1.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 17.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.8 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 4.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 4.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 4.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 6.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 8.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 5.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 4.4 | GO:0014069 | postsynaptic density(GO:0014069) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.1 | 6.7 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.9 | 4.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.9 | 5.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.8 | 2.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.7 | 2.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.5 | 1.4 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.4 | 2.9 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.4 | 3.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 1.9 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.3 | 1.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 1.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.3 | 2.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 3.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 7.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 2.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 0.8 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 1.5 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 7.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 5.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 4.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 6.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 2.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.3 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 7.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 6.4 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.8 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 1.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 6.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 2.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 7.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 2.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 3.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 2.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 3.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 4.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 4.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 2.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 4.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 1.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 5.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 3.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |