Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Hoxb8_Pdx1
Z-value: 1.35
Transcription factors associated with Hoxb8_Pdx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
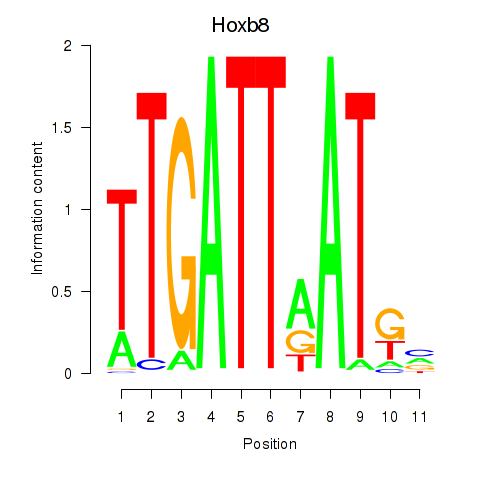
Hoxb8
|
ENSMUSG00000056648.6 | Hoxb8 |
|
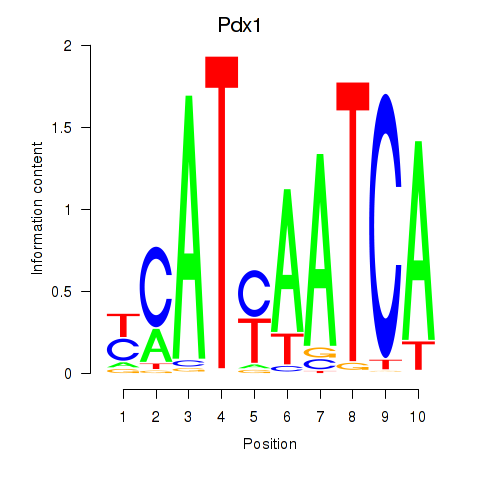
Pdx1
|
ENSMUSG00000029644.8 | Pdx1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pdx1 | mm39_v1_chr5_+_147206769_147206912 | 0.15 | 2.2e-01 | Click! |
| Hoxb8 | mm39_v1_chr11_+_96173475_96173551 | 0.02 | 8.5e-01 | Click! |
Activity profile of Hoxb8_Pdx1 motif
Sorted Z-values of Hoxb8_Pdx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb8_Pdx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_130633776 | 26.59 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr1_+_130754413 | 24.43 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr15_+_31224616 | 13.21 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr15_+_31225302 | 12.09 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chr14_+_47611270 | 11.33 |
ENSMUST00000142734.8
ENSMUST00000150290.9 ENSMUST00000226585.2 ENSMUST00000144794.2 ENSMUST00000146468.4 |
Lgals3
|
lectin, galactose binding, soluble 3 |
| chr11_-_58504307 | 11.32 |
ENSMUST00000048801.8
|
Lypd8l
|
LY6/PLAUR domain containing 8 like |
| chr13_+_4109566 | 11.22 |
ENSMUST00000041768.7
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr6_-_52141796 | 11.03 |
ENSMUST00000014848.11
|
Hoxa2
|
homeobox A2 |
| chr8_-_110765983 | 10.74 |
ENSMUST00000109222.4
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr16_-_18904240 | 10.55 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr13_-_24098981 | 9.70 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr15_+_31224460 | 9.65 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr15_+_31224555 | 9.12 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr13_-_24098951 | 9.03 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr2_-_84605764 | 8.72 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr2_-_84605732 | 8.19 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr1_-_36312482 | 7.99 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr11_+_96242422 | 7.83 |
ENSMUST00000100523.7
|
Hoxb2
|
homeobox B2 |
| chr10_+_101994719 | 7.45 |
ENSMUST00000138522.8
ENSMUST00000163753.8 ENSMUST00000138016.8 |
Mgat4c
|
MGAT4 family, member C |
| chr6_+_128639342 | 7.28 |
ENSMUST00000032518.7
ENSMUST00000204416.2 |
Clec2h
|
C-type lectin domain family 2, member h |
| chr2_+_122479770 | 7.12 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr6_+_41928559 | 6.71 |
ENSMUST00000031898.5
|
Sval1
|
seminal vesicle antigen-like 1 |
| chr10_+_101994841 | 6.41 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr1_+_88062508 | 6.41 |
ENSMUST00000113134.8
ENSMUST00000140092.8 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr1_+_88030951 | 6.24 |
ENSMUST00000113135.6
ENSMUST00000113138.8 |
Ugt1a7c
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A7C UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr19_-_4548602 | 6.23 |
ENSMUST00000048482.8
|
2010003K11Rik
|
RIKEN cDNA 2010003K11 gene |
| chr19_+_30210320 | 6.09 |
ENSMUST00000025797.7
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr4_-_35157405 | 5.46 |
ENSMUST00000102975.10
|
Mob3b
|
MOB kinase activator 3B |
| chr2_+_122478882 | 5.13 |
ENSMUST00000142767.8
|
AA467197
|
expressed sequence AA467197 |
| chr7_-_46392403 | 5.09 |
ENSMUST00000128088.4
|
Saa1
|
serum amyloid A 1 |
| chr8_-_5155347 | 5.07 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr5_+_92831150 | 4.93 |
ENSMUST00000113055.9
|
Shroom3
|
shroom family member 3 |
| chr6_-_34965339 | 4.92 |
ENSMUST00000201355.4
|
Slc23a4
|
solute carrier family 23 member 4 |
| chr19_-_24178000 | 4.88 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr13_+_94954202 | 4.85 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr8_-_4829519 | 4.78 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr2_+_125876566 | 4.78 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
| chr2_+_74512638 | 4.77 |
ENSMUST00000142312.3
|
Hoxd11
|
homeobox D11 |
| chr16_+_96162854 | 4.73 |
ENSMUST00000113794.8
|
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr3_-_20329823 | 4.72 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr5_+_92831468 | 4.61 |
ENSMUST00000168878.8
|
Shroom3
|
shroom family member 3 |
| chr13_+_4283729 | 4.48 |
ENSMUST00000081326.7
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr8_-_4829473 | 4.46 |
ENSMUST00000207262.2
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr3_-_65299967 | 4.44 |
ENSMUST00000119896.2
|
Ssr3
|
signal sequence receptor, gamma |
| chr6_-_119365632 | 4.40 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr4_-_150087587 | 4.14 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr12_-_115172211 | 4.09 |
ENSMUST00000103526.3
|
Ighv1-55
|
immunoglobulin heavy variable 1-55 |
| chr4_+_101574601 | 4.09 |
ENSMUST00000102777.10
ENSMUST00000106921.9 ENSMUST00000037552.10 ENSMUST00000145024.2 |
Lepr
|
leptin receptor |
| chr5_-_87288177 | 4.08 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr10_+_96453408 | 4.07 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr19_-_39729431 | 4.07 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr3_+_57332735 | 4.05 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr6_-_68609426 | 3.95 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr5_+_93045837 | 3.94 |
ENSMUST00000113051.9
|
Shroom3
|
shroom family member 3 |
| chr13_+_4099001 | 3.91 |
ENSMUST00000118717.10
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr2_+_125876883 | 3.89 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7 |
| chr6_-_136758716 | 3.86 |
ENSMUST00000078095.11
ENSMUST00000032338.10 |
Gucy2c
|
guanylate cyclase 2c |
| chr4_+_148985584 | 3.84 |
ENSMUST00000147270.2
|
Casz1
|
castor zinc finger 1 |
| chr6_-_3399451 | 3.83 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr6_-_124519240 | 3.80 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr3_-_65300000 | 3.76 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chr18_-_32271224 | 3.71 |
ENSMUST00000234657.2
ENSMUST00000234386.2 ENSMUST00000234651.2 |
Proc
|
protein C |
| chr11_-_84058292 | 3.56 |
ENSMUST00000050771.8
|
Gm11437
|
predicted gene 11437 |
| chr5_-_88675700 | 3.50 |
ENSMUST00000087033.6
|
Jchain
|
immunoglobulin joining chain |
| chr5_-_113369096 | 3.42 |
ENSMUST00000211733.2
|
2900026A02Rik
|
RIKEN cDNA 2900026A02 gene |
| chr1_+_88093726 | 3.35 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr12_-_115722081 | 3.28 |
ENSMUST00000103541.3
|
Ighv1-72
|
immunoglobulin heavy variable 1-72 |
| chr9_-_70842090 | 3.25 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr9_-_31043076 | 3.22 |
ENSMUST00000034478.3
|
St14
|
suppression of tumorigenicity 14 (colon carcinoma) |
| chr15_+_101310283 | 3.21 |
ENSMUST00000068904.9
|
Krt7
|
keratin 7 |
| chr3_-_157630690 | 3.15 |
ENSMUST00000118539.2
|
Cth
|
cystathionase (cystathionine gamma-lyase) |
| chr3_+_132335704 | 3.12 |
ENSMUST00000212594.2
|
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr8_-_110766009 | 3.09 |
ENSMUST00000212934.2
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr3_-_88417251 | 3.07 |
ENSMUST00000149068.2
|
Lmna
|
lamin A |
| chr16_+_22769822 | 3.07 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr6_+_116627567 | 3.04 |
ENSMUST00000067354.10
ENSMUST00000178241.4 |
Depp1
|
DEPP1 autophagy regulator |
| chr16_-_19079594 | 3.04 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr9_-_76474374 | 3.04 |
ENSMUST00000183437.8
|
Fam83b
|
family with sequence similarity 83, member B |
| chr6_-_52203146 | 3.04 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr2_-_74489763 | 3.03 |
ENSMUST00000173623.2
ENSMUST00000001867.13 |
Evx2
|
even-skipped homeobox 2 |
| chr17_-_41225241 | 3.03 |
ENSMUST00000166343.3
|
Glyatl3
|
glycine-N-acyltransferase-like 3 |
| chr6_-_52185674 | 3.01 |
ENSMUST00000062829.9
|
Hoxa6
|
homeobox A6 |
| chr3_-_107851021 | 2.99 |
ENSMUST00000106684.8
ENSMUST00000106685.9 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr12_-_103923145 | 2.97 |
ENSMUST00000085054.5
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr12_-_103829810 | 2.95 |
ENSMUST00000085056.8
ENSMUST00000072876.12 ENSMUST00000124717.2 |
Serpina1a
|
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
| chr1_-_45542442 | 2.92 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr17_+_37504783 | 2.91 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr3_-_82957104 | 2.82 |
ENSMUST00000048246.5
|
Fgb
|
fibrinogen beta chain |
| chr13_+_19398273 | 2.75 |
ENSMUST00000103558.3
|
Trgc1
|
T cell receptor gamma, constant 1 |
| chr6_+_142244145 | 2.63 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr9_-_70841881 | 2.62 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chr9_-_85209340 | 2.61 |
ENSMUST00000187711.2
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr14_+_74973081 | 2.59 |
ENSMUST00000177283.8
|
Esd
|
esterase D/formylglutathione hydrolase |
| chr9_-_85209162 | 2.58 |
ENSMUST00000034802.15
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr6_+_124281607 | 2.56 |
ENSMUST00000032234.5
ENSMUST00000112541.8 |
Cd163
|
CD163 antigen |
| chr16_+_78727829 | 2.56 |
ENSMUST00000114216.2
ENSMUST00000069148.13 ENSMUST00000023568.14 |
Chodl
|
chondrolectin |
| chr1_+_87983099 | 2.54 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr3_-_107850707 | 2.54 |
ENSMUST00000106681.3
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr5_-_130053120 | 2.52 |
ENSMUST00000161640.8
ENSMUST00000161884.2 ENSMUST00000161094.8 |
Asl
|
argininosuccinate lyase |
| chr11_-_69439934 | 2.51 |
ENSMUST00000108659.2
|
Dnah2
|
dynein, axonemal, heavy chain 2 |
| chr5_-_87074380 | 2.50 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr1_-_57011595 | 2.49 |
ENSMUST00000042857.14
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr1_-_10038030 | 2.46 |
ENSMUST00000185184.2
|
Tcf24
|
transcription factor 24 |
| chr6_-_3494587 | 2.45 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr3_+_132335575 | 2.44 |
ENSMUST00000212804.2
ENSMUST00000212852.2 |
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr1_-_39616445 | 2.44 |
ENSMUST00000062525.11
|
Rnf149
|
ring finger protein 149 |
| chr17_-_71153283 | 2.42 |
ENSMUST00000156484.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr10_+_5543769 | 2.40 |
ENSMUST00000051809.10
|
Myct1
|
myc target 1 |
| chr13_+_23991010 | 2.39 |
ENSMUST00000006786.11
ENSMUST00000099697.3 |
Slc17a2
|
solute carrier family 17 (sodium phosphate), member 2 |
| chr16_+_22769844 | 2.35 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr13_+_94219934 | 2.33 |
ENSMUST00000156071.2
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr7_+_46401214 | 2.32 |
ENSMUST00000210769.2
ENSMUST00000210272.2 ENSMUST00000075982.4 |
Saa2
|
serum amyloid A 2 |
| chr17_-_71309815 | 2.31 |
ENSMUST00000123686.8
|
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr16_-_56533179 | 2.31 |
ENSMUST00000136394.8
|
Tfg
|
Trk-fused gene |
| chr2_+_152873772 | 2.31 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr17_-_35081129 | 2.27 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr5_+_30360246 | 2.27 |
ENSMUST00000026841.15
ENSMUST00000123980.8 ENSMUST00000114783.6 ENSMUST00000114786.8 |
Hadhb
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
| chr5_-_30360113 | 2.26 |
ENSMUST00000156859.3
|
Hadha
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha |
| chr1_-_120198804 | 2.23 |
ENSMUST00000112641.8
|
Steap3
|
STEAP family member 3 |
| chr11_-_99482165 | 2.22 |
ENSMUST00000104930.2
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr2_+_69210775 | 2.20 |
ENSMUST00000063690.4
|
Dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr1_-_126758369 | 2.19 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr18_-_66155651 | 2.18 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr2_-_69036489 | 2.18 |
ENSMUST00000127243.8
ENSMUST00000149643.2 ENSMUST00000167875.9 ENSMUST00000005365.15 |
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr11_+_96214078 | 2.17 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr5_-_87572060 | 2.16 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr12_-_115425105 | 2.14 |
ENSMUST00000103532.3
|
Ighv1-62-3
|
immunoglobulin heavy variable 1-62-3 |
| chr3_-_51184730 | 2.14 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr3_+_85948030 | 2.11 |
ENSMUST00000238545.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr10_+_101517348 | 2.06 |
ENSMUST00000179929.8
ENSMUST00000219195.2 ENSMUST00000127504.9 |
Mgat4c
|
MGAT4 family, member C |
| chr3_+_82915031 | 2.05 |
ENSMUST00000048486.13
ENSMUST00000194175.2 |
Fgg
|
fibrinogen gamma chain |
| chr18_+_43897354 | 2.03 |
ENSMUST00000187157.7
ENSMUST00000043803.13 ENSMUST00000189750.2 |
Scgb3a2
|
secretoglobin, family 3A, member 2 |
| chr2_-_38604503 | 2.01 |
ENSMUST00000028084.5
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr19_+_39980868 | 2.00 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr5_-_66211842 | 2.00 |
ENSMUST00000200852.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr3_+_144824644 | 1.99 |
ENSMUST00000199124.5
|
Odf2l
|
outer dense fiber of sperm tails 2-like |
| chr1_-_149798090 | 1.95 |
ENSMUST00000111926.9
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr6_-_21851827 | 1.92 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr3_+_85947806 | 1.88 |
ENSMUST00000238222.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr1_-_164763091 | 1.87 |
ENSMUST00000027860.8
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr2_-_155668567 | 1.87 |
ENSMUST00000109638.2
ENSMUST00000134278.2 |
Eif6
|
eukaryotic translation initiation factor 6 |
| chr17_+_79919267 | 1.84 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr17_-_35081456 | 1.83 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr2_-_69036472 | 1.83 |
ENSMUST00000112320.8
|
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr4_+_109092610 | 1.83 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr18_+_46983105 | 1.81 |
ENSMUST00000025358.4
ENSMUST00000234519.2 |
Lvrn
|
laeverin |
| chr14_-_59602882 | 1.81 |
ENSMUST00000160425.8
ENSMUST00000095157.11 |
Phf11d
|
PHD finger protein 11D |
| chr8_-_45747883 | 1.79 |
ENSMUST00000026907.6
|
Klkb1
|
kallikrein B, plasma 1 |
| chrX_-_105055486 | 1.77 |
ENSMUST00000238718.2
ENSMUST00000033583.14 ENSMUST00000151689.9 |
Magt1
|
magnesium transporter 1 |
| chr1_+_87983189 | 1.77 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr6_+_116627635 | 1.75 |
ENSMUST00000204555.2
|
Depp1
|
DEPP1 autophagy regulator |
| chr14_+_55797468 | 1.75 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr1_-_57010921 | 1.73 |
ENSMUST00000114415.10
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr7_-_37472979 | 1.72 |
ENSMUST00000176534.8
|
Zfp536
|
zinc finger protein 536 |
| chr1_-_126758520 | 1.71 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr14_+_26722319 | 1.68 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr1_+_118555668 | 1.65 |
ENSMUST00000027629.10
|
Tfcp2l1
|
transcription factor CP2-like 1 |
| chr10_+_33733706 | 1.65 |
ENSMUST00000218204.2
|
Sult3a1
|
sulfotransferase family 3A, member 1 |
| chr19_-_32080496 | 1.64 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr9_+_35334878 | 1.62 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr1_-_162726053 | 1.62 |
ENSMUST00000143123.3
|
Fmo2
|
flavin containing monooxygenase 2 |
| chr8_-_84059048 | 1.61 |
ENSMUST00000177594.8
ENSMUST00000053902.4 |
Elmod2
|
ELMO/CED-12 domain containing 2 |
| chr10_+_100428212 | 1.61 |
ENSMUST00000187119.7
ENSMUST00000188736.7 ENSMUST00000191336.7 |
1700017N19Rik
|
RIKEN cDNA 1700017N19 gene |
| chr8_-_106198112 | 1.60 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr14_+_55797443 | 1.55 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr2_-_84255602 | 1.55 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr1_+_58752415 | 1.54 |
ENSMUST00000114309.8
ENSMUST00000069333.8 |
Cflar
|
CASP8 and FADD-like apoptosis regulator |
| chr5_+_65920861 | 1.49 |
ENSMUST00000201489.4
|
N4bp2
|
NEDD4 binding protein 2 |
| chr18_-_35631914 | 1.49 |
ENSMUST00000236007.2
ENSMUST00000237896.2 ENSMUST00000235778.2 ENSMUST00000235524.2 ENSMUST00000235691.2 ENSMUST00000235619.2 ENSMUST00000025215.10 |
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr2_+_125701054 | 1.48 |
ENSMUST00000028636.13
ENSMUST00000125084.8 |
Galk2
|
galactokinase 2 |
| chr16_+_90535212 | 1.42 |
ENSMUST00000038197.3
|
Mrap
|
melanocortin 2 receptor accessory protein |
| chr14_+_55797934 | 1.41 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr14_+_54433524 | 1.40 |
ENSMUST00000103718.2
|
Traj23
|
T cell receptor alpha joining 23 |
| chr1_-_65071897 | 1.38 |
ENSMUST00000162800.8
ENSMUST00000069142.12 |
Akr1cl
|
aldo-keto reductase family 1, member C-like |
| chr5_-_125418107 | 1.38 |
ENSMUST00000111390.8
ENSMUST00000086075.13 |
Scarb1
|
scavenger receptor class B, member 1 |
| chr7_-_49286594 | 1.38 |
ENSMUST00000032717.7
|
Dbx1
|
developing brain homeobox 1 |
| chr10_-_128579879 | 1.38 |
ENSMUST00000026414.9
|
Dgka
|
diacylglycerol kinase, alpha |
| chr1_+_90981144 | 1.36 |
ENSMUST00000068167.13
ENSMUST00000097649.10 ENSMUST00000186762.7 ENSMUST00000097650.10 |
Lrrfip1
|
leucine rich repeat (in FLII) interacting protein 1 |
| chrM_+_11735 | 1.35 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr12_-_103871146 | 1.35 |
ENSMUST00000074051.6
|
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1C |
| chr1_-_118981069 | 1.32 |
ENSMUST00000161301.8
ENSMUST00000161451.8 ENSMUST00000162607.2 |
Gli2
|
GLI-Kruppel family member GLI2 |
| chr17_+_24645615 | 1.32 |
ENSMUST00000234304.2
ENSMUST00000024946.7 ENSMUST00000234150.2 |
Eci1
|
enoyl-Coenzyme A delta isomerase 1 |
| chr9_+_54441404 | 1.30 |
ENSMUST00000118413.3
|
Sh2d7
|
SH2 domain containing 7 |
| chr16_+_24540599 | 1.30 |
ENSMUST00000115314.4
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr17_-_35023521 | 1.29 |
ENSMUST00000025223.9
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr6_-_51989456 | 1.29 |
ENSMUST00000078214.8
ENSMUST00000204778.3 |
Skap2
|
src family associated phosphoprotein 2 |
| chr19_-_3625698 | 1.27 |
ENSMUST00000172362.3
ENSMUST00000025846.16 ENSMUST00000226109.2 ENSMUST00000113997.9 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr19_+_11674413 | 1.27 |
ENSMUST00000069760.13
ENSMUST00000119053.3 |
Oosp3
|
oocyte secreted protein 3 |
| chr10_-_41585293 | 1.26 |
ENSMUST00000019955.16
ENSMUST00000099932.10 |
Ccdc162
|
coiled-coil domain containing 162 |
| chr3_-_51184895 | 1.25 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr2_-_125701059 | 1.25 |
ENSMUST00000110463.8
ENSMUST00000028635.6 |
Cops2
|
COP9 signalosome subunit 2 |
| chr11_-_106890307 | 1.22 |
ENSMUST00000018506.13
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr8_-_25085654 | 1.19 |
ENSMUST00000110667.8
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr1_-_37575313 | 1.19 |
ENSMUST00000042161.15
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr4_+_138503046 | 1.18 |
ENSMUST00000030528.9
|
Pla2g2d
|
phospholipase A2, group IID |
| chr2_+_36120438 | 1.18 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr11_+_46701619 | 1.17 |
ENSMUST00000068877.7
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr12_+_105569344 | 1.16 |
ENSMUST00000182899.2
ENSMUST00000183086.2 |
Bdkrb1
|
bradykinin receptor, beta 1 |
| chr14_+_54429757 | 1.16 |
ENSMUST00000103714.2
|
Traj27
|
T cell receptor alpha joining 27 |
| chr12_-_16850887 | 1.15 |
ENSMUST00000161998.3
|
Greb1
|
gene regulated by estrogen in breast cancer protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 24.4 | GO:0002414 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 5.6 | 16.9 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 4.6 | 13.8 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 3.1 | 18.9 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 2.9 | 8.7 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 2.2 | 8.9 | GO:2001189 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 2.1 | 6.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 1.8 | 5.4 | GO:0097037 | heme export(GO:0097037) |
| 1.4 | 14.1 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 1.3 | 3.8 | GO:0060220 | camera-type eye photoreceptor cell fate commitment(GO:0060220) negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 1.3 | 2.5 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.2 | 5.9 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 1.1 | 3.2 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 1.0 | 4.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 1.0 | 26.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 1.0 | 9.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.0 | 4.8 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.9 | 2.6 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.8 | 2.5 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.8 | 3.1 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.7 | 2.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.7 | 2.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.7 | 44.1 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.7 | 4.9 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.6 | 1.9 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) positive regulation of thymocyte migration(GO:2000412) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.6 | 1.9 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.6 | 2.9 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.5 | 8.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.5 | 8.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.5 | 3.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.5 | 1.4 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.5 | 4.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.4 | 1.3 | GO:0060032 | floor plate formation(GO:0021508) notochord regression(GO:0060032) |
| 0.4 | 3.7 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.4 | 1.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.4 | 3.5 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.4 | 1.5 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.4 | 1.5 | GO:1903943 | skeletal muscle atrophy(GO:0014732) negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.4 | 3.8 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.4 | 3.3 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 2.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.4 | 1.8 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.3 | 1.0 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.3 | 3.7 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.3 | 2.6 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.3 | 4.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.3 | 2.0 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.3 | 4.8 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.3 | 2.0 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 1.6 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.3 | 2.7 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 1.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.2 | 4.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 2.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 4.1 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.2 | 1.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 1.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.2 | 0.8 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.2 | 2.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 6.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 0.5 | GO:0016131 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.2 | 4.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 1.9 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.2 | 1.0 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.2 | 0.5 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.2 | 0.5 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.2 | 5.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 2.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.9 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.9 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 0.4 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 13.7 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 | 3.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 1.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 1.7 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.1 | 1.0 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 1.3 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 1.0 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.6 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 6.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 1.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.7 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 1.0 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.1 | 0.5 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 1.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 1.4 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 1.2 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 | 1.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 3.0 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.8 | GO:0019661 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 | 1.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 1.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 14.0 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 1.3 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) |
| 0.1 | 0.3 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 4.2 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 0.2 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.1 | 0.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.5 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.6 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 10.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 6.9 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 0.5 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.8 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.8 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 1.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.3 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 1.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.7 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 5.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.6 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.8 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 0.7 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 0.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.2 | GO:0035938 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.8 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.6 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 1.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 1.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 4.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.2 | GO:1904171 | negative regulation of bleb assembly(GO:1904171) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 9.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 1.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.7 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.3 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.5 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.7 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.0 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 3.3 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.4 | GO:2000269 | fibroblast apoptotic process(GO:0044346) regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 | 0.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.1 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 2.4 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.2 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 1.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 2.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.6 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 1.4 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 12.6 | GO:0005975 | carbohydrate metabolic process(GO:0005975) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 1.0 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 1.3 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.3 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.3 | GO:0019724 | B cell mediated immunity(GO:0019724) |
| 0.0 | 0.4 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:0060459 | left lung development(GO:0060459) left lung morphogenesis(GO:0060460) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 32.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 1.0 | 2.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 4.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.7 | 4.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.6 | 12.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.5 | 4.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 22.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 13.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 11.3 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.3 | 1.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 3.0 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 2.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 1.3 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 1.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 4.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 40.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 5.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 2.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.0 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 1.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 4.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.3 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.1 | 1.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 2.2 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 0.4 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 4.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 7.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.0 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 2.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 2.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 1.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 12.4 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 2.3 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 2.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 4.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 3.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 6.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 39.1 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 13.8 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 26.6 | GO:0035375 | zymogen binding(GO:0035375) |
| 3.8 | 11.3 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 2.5 | 15.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 2.1 | 44.1 | GO:0070513 | death domain binding(GO:0070513) |
| 2.0 | 24.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 2.0 | 13.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.6 | 18.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 1.5 | 4.5 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 1.4 | 4.1 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 1.3 | 21.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.2 | 8.7 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 1.2 | 5.9 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.9 | 7.6 | GO:0005534 | galactose binding(GO:0005534) |
| 0.9 | 4.4 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.9 | 3.5 | GO:0019862 | IgA binding(GO:0019862) |
| 0.8 | 3.0 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.7 | 3.7 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.7 | 29.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.6 | 5.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.6 | 2.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.6 | 16.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.5 | 1.5 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.5 | 1.4 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.4 | 12.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 1.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.4 | 1.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.4 | 2.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.4 | 7.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.4 | 2.9 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 1.4 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.3 | 4.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.3 | 1.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 1.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.3 | 2.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 1.5 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.2 | 2.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.7 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.2 | 7.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 2.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 1.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 1.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 0.8 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.2 | 3.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 0.5 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.2 | 6.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 4.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 4.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 0.8 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 5.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 6.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 5.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 2.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.0 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 1.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 2.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 4.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.6 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 3.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 2.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.6 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 1.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 2.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 10.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.4 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.1 | 3.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.1 | 0.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 10.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 3.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 12.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.8 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 1.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 1.9 | GO:0008009 | chemokine activity(GO:0008009) CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 12.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 2.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 11.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 5.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 2.1 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 3.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 1.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 17.2 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 24.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 11.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 4.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 5.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 27.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 26.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 5.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 3.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 3.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 4.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 3.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 5.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 2.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 4.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 18.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.8 | 6.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.7 | 16.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.7 | 18.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.6 | 8.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.5 | 4.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.5 | 5.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 5.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 3.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 4.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 7.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.2 | 4.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 5.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 3.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 3.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 4.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 10.0 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 1.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 2.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 3.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 4.6 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.1 | 1.9 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 4.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 3.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.6 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |