Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
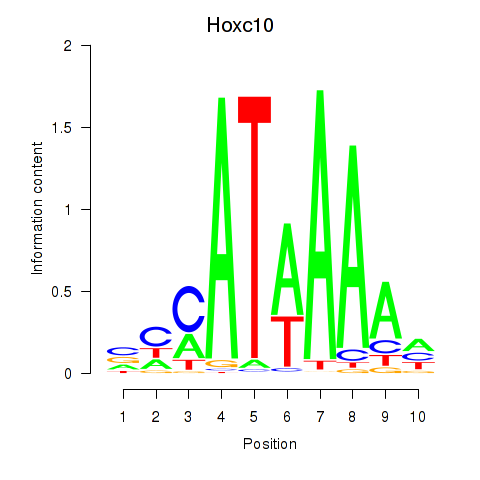
Results for Hoxc10
Z-value: 0.93
Transcription factors associated with Hoxc10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc10
|
ENSMUSG00000022484.8 | Hoxc10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc10 | mm39_v1_chr15_+_102875229_102875248 | -0.36 | 2.2e-03 | Click! |
Activity profile of Hoxc10 motif
Sorted Z-values of Hoxc10 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc10
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_31363245 | 8.71 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr3_+_69129745 | 7.06 |
ENSMUST00000183126.2
|
Arl14
|
ADP-ribosylation factor-like 14 |
| chr1_-_45542442 | 6.84 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr3_-_63758672 | 6.80 |
ENSMUST00000162269.9
ENSMUST00000159676.9 ENSMUST00000175947.8 |
Plch1
|
phospholipase C, eta 1 |
| chr8_-_62576140 | 6.69 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr11_-_100036792 | 6.63 |
ENSMUST00000007317.8
|
Krt19
|
keratin 19 |
| chr1_-_149836974 | 4.66 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr18_-_70274639 | 4.53 |
ENSMUST00000121693.8
|
Rab27b
|
RAB27B, member RAS oncogene family |
| chr17_+_31652073 | 3.59 |
ENSMUST00000237363.2
|
Pde9a
|
phosphodiesterase 9A |
| chr9_+_75682637 | 3.36 |
ENSMUST00000012281.8
|
Bmp5
|
bone morphogenetic protein 5 |
| chr17_+_31652029 | 3.30 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr16_-_38620688 | 3.21 |
ENSMUST00000057767.6
|
Upk1b
|
uroplakin 1B |
| chr7_-_129867967 | 3.09 |
ENSMUST00000117872.8
ENSMUST00000120187.9 |
Fgfr2
|
fibroblast growth factor receptor 2 |
| chr19_+_39102342 | 3.07 |
ENSMUST00000087234.3
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr1_-_160079007 | 3.00 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr16_-_55659194 | 2.81 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr7_-_129867921 | 2.81 |
ENSMUST00000122054.8
|
Fgfr2
|
fibroblast growth factor receptor 2 |
| chr17_-_35293447 | 2.64 |
ENSMUST00000007259.4
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr11_-_79394904 | 2.36 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr16_+_41353360 | 2.33 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr11_-_83957889 | 2.33 |
ENSMUST00000108101.8
|
Dusp14
|
dual specificity phosphatase 14 |
| chr11_-_43492367 | 2.28 |
ENSMUST00000020672.5
|
Fabp6
|
fatty acid binding protein 6 |
| chr11_-_97591150 | 2.28 |
ENSMUST00000018681.14
|
Pcgf2
|
polycomb group ring finger 2 |
| chr16_+_29884153 | 2.21 |
ENSMUST00000023171.8
|
Hes1
|
hes family bHLH transcription factor 1 |
| chrX_+_138464065 | 2.11 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr10_+_90412827 | 1.97 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr15_-_101441254 | 1.94 |
ENSMUST00000023720.8
|
Krt84
|
keratin 84 |
| chr9_+_21914083 | 1.93 |
ENSMUST00000216344.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr7_-_30312246 | 1.92 |
ENSMUST00000006476.6
|
Upk1a
|
uroplakin 1A |
| chrX_+_95498965 | 1.89 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr1_-_144124871 | 1.89 |
ENSMUST00000189061.7
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr7_+_43339842 | 1.83 |
ENSMUST00000056329.7
|
Klk14
|
kallikrein related-peptidase 14 |
| chr1_-_182109773 | 1.80 |
ENSMUST00000133052.2
|
Degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr3_+_3699205 | 1.79 |
ENSMUST00000108394.3
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr15_-_101336669 | 1.72 |
ENSMUST00000081945.5
|
Krt87
|
keratin 87 |
| chr3_+_63988968 | 1.71 |
ENSMUST00000029406.6
|
Vmn2r1
|
vomeronasal 2, receptor 1 |
| chr11_+_100902572 | 1.59 |
ENSMUST00000092663.4
|
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr12_-_69838004 | 1.58 |
ENSMUST00000221646.2
|
Cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr10_-_6930376 | 1.47 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr7_-_30750856 | 1.47 |
ENSMUST00000073892.6
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr14_+_27598021 | 1.46 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr1_+_66426127 | 1.41 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr12_+_117621644 | 1.40 |
ENSMUST00000222105.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr1_+_107439145 | 1.37 |
ENSMUST00000009356.11
ENSMUST00000064916.9 |
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr2_+_3115250 | 1.31 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr11_+_96085118 | 1.31 |
ENSMUST00000062709.4
|
Hoxb13
|
homeobox B13 |
| chr3_-_146475974 | 1.28 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr19_+_56536685 | 1.24 |
ENSMUST00000071423.7
|
Nhlrc2
|
NHL repeat containing 2 |
| chr6_-_136852792 | 1.24 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr7_-_30750828 | 1.23 |
ENSMUST00000206341.2
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr3_+_89173859 | 1.22 |
ENSMUST00000040824.2
|
Dpm3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr1_-_22551594 | 1.17 |
ENSMUST00000239255.2
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr18_-_43032359 | 1.14 |
ENSMUST00000117687.8
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr9_-_116004386 | 1.13 |
ENSMUST00000035014.8
|
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr19_-_7319157 | 1.13 |
ENSMUST00000164205.8
ENSMUST00000165286.8 ENSMUST00000168324.2 ENSMUST00000032557.15 |
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr9_+_53678801 | 1.11 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr1_+_179788675 | 1.09 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_+_83642910 | 1.09 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr8_+_94537910 | 1.08 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr5_+_43672856 | 1.08 |
ENSMUST00000076939.10
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr19_-_56536443 | 1.06 |
ENSMUST00000182059.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr1_-_86039692 | 1.05 |
ENSMUST00000027431.7
|
Htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B |
| chr3_-_117153802 | 1.02 |
ENSMUST00000197743.2
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr19_-_56536646 | 1.01 |
ENSMUST00000182276.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr10_+_69816153 | 1.01 |
ENSMUST00000182207.8
|
Ank3
|
ankyrin 3, epithelial |
| chr17_-_34043502 | 1.00 |
ENSMUST00000087342.13
ENSMUST00000173844.8 |
Rps28
|
ribosomal protein S28 |
| chr10_-_49659355 | 0.99 |
ENSMUST00000105484.10
ENSMUST00000218598.2 ENSMUST00000079751.9 ENSMUST00000218441.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr2_+_163348728 | 0.98 |
ENSMUST00000143911.8
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chrX_+_55493325 | 0.98 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr11_+_115824108 | 0.96 |
ENSMUST00000140991.2
|
Sap30bp
|
SAP30 binding protein |
| chr2_+_80145805 | 0.95 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr3_-_49711706 | 0.94 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr19_-_7319211 | 0.91 |
ENSMUST00000171393.8
|
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr2_-_32278245 | 0.90 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr1_-_144124787 | 0.89 |
ENSMUST00000185714.2
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr14_+_54923655 | 0.87 |
ENSMUST00000038539.8
ENSMUST00000228027.2 |
1700123O20Rik
|
RIKEN cDNA 1700123O20 gene |
| chr3_+_31956814 | 0.81 |
ENSMUST00000192429.6
ENSMUST00000191869.6 ENSMUST00000178668.2 ENSMUST00000119970.8 |
Kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr17_-_90395771 | 0.80 |
ENSMUST00000197268.5
ENSMUST00000173917.8 |
Nrxn1
|
neurexin I |
| chr17_-_34043320 | 0.80 |
ENSMUST00000173879.8
ENSMUST00000166693.3 ENSMUST00000173019.8 |
Rps28
|
ribosomal protein S28 |
| chr2_+_113235475 | 0.77 |
ENSMUST00000238883.2
|
Fmn1
|
formin 1 |
| chr2_-_32277773 | 0.76 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr2_+_65499097 | 0.73 |
ENSMUST00000200829.4
|
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr12_-_113912416 | 0.71 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chr3_-_49711765 | 0.70 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr12_+_59142439 | 0.70 |
ENSMUST00000219140.3
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr13_+_25127127 | 0.69 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr5_+_71857261 | 0.68 |
ENSMUST00000031122.9
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 |
| chr15_+_25774070 | 0.68 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr11_+_96214078 | 0.67 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr11_+_108811168 | 0.65 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr15_+_6451721 | 0.65 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chrX_+_132809189 | 0.65 |
ENSMUST00000113304.2
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr14_-_9015639 | 0.65 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chr12_+_105570350 | 0.65 |
ENSMUST00000041229.5
|
Bdkrb1
|
bradykinin receptor, beta 1 |
| chr2_-_86180622 | 0.64 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr5_-_62923463 | 0.64 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr9_-_44473146 | 0.64 |
ENSMUST00000215293.2
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr4_+_11758147 | 0.62 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr4_+_102111936 | 0.61 |
ENSMUST00000106907.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr13_+_38223023 | 0.59 |
ENSMUST00000226110.2
|
Riok1
|
RIO kinase 1 |
| chr4_-_58787716 | 0.58 |
ENSMUST00000216719.2
|
Olfr267
|
olfactory receptor 267 |
| chr1_+_189460461 | 0.58 |
ENSMUST00000097442.9
|
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr18_-_43032535 | 0.57 |
ENSMUST00000120632.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chrX_+_168468186 | 0.51 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr11_-_115824290 | 0.50 |
ENSMUST00000021097.10
|
Recql5
|
RecQ protein-like 5 |
| chr13_+_104424359 | 0.49 |
ENSMUST00000065766.7
|
Adamts6
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 6 |
| chr2_+_87687521 | 0.48 |
ENSMUST00000061081.2
|
Olfr1151
|
olfactory receptor 1151 |
| chr5_+_43673093 | 0.48 |
ENSMUST00000144558.3
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr15_-_101389384 | 0.46 |
ENSMUST00000023718.9
|
Krt83
|
keratin 83 |
| chr19_-_50667079 | 0.46 |
ENSMUST00000209413.2
ENSMUST00000072685.13 ENSMUST00000164039.9 |
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr2_+_73102269 | 0.45 |
ENSMUST00000090813.6
|
Sp9
|
trans-acting transcription factor 9 |
| chr11_+_108811626 | 0.44 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr11_-_102210568 | 0.43 |
ENSMUST00000173870.8
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr15_+_91722524 | 0.42 |
ENSMUST00000109276.8
ENSMUST00000088555.10 ENSMUST00000100293.9 ENSMUST00000126508.8 ENSMUST00000239545.1 |
Smgc
ENSMUSG00000118670.1
|
submandibular gland protein C mucin 19 |
| chr4_-_154721288 | 0.40 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr15_-_101422054 | 0.40 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chr14_-_54923517 | 0.40 |
ENSMUST00000125265.2
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr17_-_37523969 | 0.40 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr1_+_17672117 | 0.39 |
ENSMUST00000088476.4
|
Pi15
|
peptidase inhibitor 15 |
| chr10_+_129153986 | 0.39 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr7_-_29605504 | 0.38 |
ENSMUST00000053521.15
|
Zfp27
|
zinc finger protein 27 |
| chr6_+_29694181 | 0.38 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr5_-_67256578 | 0.38 |
ENSMUST00000012664.11
|
Phox2b
|
paired-like homeobox 2b |
| chr7_+_108143734 | 0.37 |
ENSMUST00000078162.4
ENSMUST00000211693.2 |
Olfr503
|
olfactory receptor 503 |
| chr14_+_66043281 | 0.36 |
ENSMUST00000022612.10
|
Pbk
|
PDZ binding kinase |
| chr5_+_128677863 | 0.34 |
ENSMUST00000117102.4
|
Fzd10
|
frizzled class receptor 10 |
| chr2_-_129213050 | 0.34 |
ENSMUST00000028881.14
|
Il1b
|
interleukin 1 beta |
| chr10_+_94350687 | 0.33 |
ENSMUST00000065060.12
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr2_+_89804937 | 0.33 |
ENSMUST00000214630.2
ENSMUST00000111512.10 ENSMUST00000144710.3 ENSMUST00000216678.2 |
Olfr1260
|
olfactory receptor 1260 |
| chr19_+_55882942 | 0.33 |
ENSMUST00000142291.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr2_+_36958319 | 0.32 |
ENSMUST00000120704.2
|
Olfr360
|
olfactory receptor 360 |
| chr18_+_44249254 | 0.32 |
ENSMUST00000212114.2
|
Gm37797
|
predicted gene, 37797 |
| chr1_+_179788037 | 0.30 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr6_-_133014064 | 0.28 |
ENSMUST00000032317.4
|
Tas2r103
|
taste receptor, type 2, member 103 |
| chr1_-_36283326 | 0.28 |
ENSMUST00000046875.14
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr11_-_24025054 | 0.24 |
ENSMUST00000068360.2
|
A830031A19Rik
|
RIKEN cDNA A830031A19 gene |
| chr8_-_18791557 | 0.22 |
ENSMUST00000033846.7
|
Angpt2
|
angiopoietin 2 |
| chr2_+_177783713 | 0.21 |
ENSMUST00000103066.10
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr7_-_73187369 | 0.20 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr10_-_12689345 | 0.20 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr18_-_43032514 | 0.19 |
ENSMUST00000236238.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr6_-_24168082 | 0.15 |
ENSMUST00000031713.9
|
Slc13a1
|
solute carrier family 13 (sodium/sulfate symporters), member 1 |
| chr4_-_110148081 | 0.14 |
ENSMUST00000142722.2
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr8_-_112718888 | 0.13 |
ENSMUST00000034427.12
ENSMUST00000139820.8 |
Adat1
|
adenosine deaminase, tRNA-specific 1 |
| chr17_+_37670473 | 0.13 |
ENSMUST00000178766.3
ENSMUST00000215398.2 |
Olfr104-ps
|
olfactory receptor 104, pseudogene |
| chr7_-_10009278 | 0.12 |
ENSMUST00000060374.5
|
Vmn1r66
|
vomeronasal 1 receptor 66 |
| chr10_-_89270554 | 0.12 |
ENSMUST00000220071.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chrX_-_49108236 | 0.10 |
ENSMUST00000213556.3
ENSMUST00000213463.2 |
Olfr1323
|
olfactory receptor 1323 |
| chr10_+_87926932 | 0.10 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr15_-_78947038 | 0.10 |
ENSMUST00000151889.8
ENSMUST00000040676.11 |
Ankrd54
|
ankyrin repeat domain 54 |
| chr10_-_129023288 | 0.09 |
ENSMUST00000072976.4
|
Olfr773
|
olfactory receptor 773 |
| chr5_+_143864993 | 0.08 |
ENSMUST00000172367.3
|
Gm42421
|
predicted gene, 42421 |
| chrX_-_166907286 | 0.07 |
ENSMUST00000239138.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr10_-_129951252 | 0.05 |
ENSMUST00000213145.2
|
Olfr823
|
olfactory receptor 823 |
| chr14_-_50479323 | 0.05 |
ENSMUST00000214152.2
|
Olfr731
|
olfactory receptor 731 |
| chr19_+_26725589 | 0.05 |
ENSMUST00000207812.2
ENSMUST00000175791.9 ENSMUST00000207118.2 ENSMUST00000209085.2 ENSMUST00000112637.10 ENSMUST00000207054.2 ENSMUST00000208589.2 ENSMUST00000176475.9 ENSMUST00000176698.9 ENSMUST00000207832.2 ENSMUST00000177252.9 ENSMUST00000208712.2 ENSMUST00000208186.2 ENSMUST00000208806.2 ENSMUST00000208027.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr11_-_58614840 | 0.04 |
ENSMUST00000214728.2
|
Olfr318
|
olfactory receptor 318 |
| chr7_-_139162706 | 0.04 |
ENSMUST00000106095.3
|
Nkx6-2
|
NK6 homeobox 2 |
| chr1_-_97689263 | 0.04 |
ENSMUST00000171129.8
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr11_-_99457456 | 0.04 |
ENSMUST00000055502.5
|
Krtap3-1
|
keratin associated protein 3-1 |
| chr2_+_89821565 | 0.03 |
ENSMUST00000111509.3
ENSMUST00000213909.3 |
Olfr1261
|
olfactory receptor 1261 |
| chr16_-_19425453 | 0.02 |
ENSMUST00000078603.3
ENSMUST00000218837.2 |
Olfr170
|
olfactory receptor 170 |
| chr11_-_73215442 | 0.02 |
ENSMUST00000021119.9
|
Aspa
|
aspartoacylase |
| chr1_-_63153414 | 0.02 |
ENSMUST00000153992.2
ENSMUST00000165066.8 ENSMUST00000172416.8 ENSMUST00000137511.8 |
Ino80d
|
INO80 complex subunit D |
| chr17_+_37689924 | 0.01 |
ENSMUST00000215518.2
|
Olfr105-ps
|
olfactory receptor 105, pseudogene |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) |
| 1.7 | 6.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.2 | 8.7 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 1.1 | 2.2 | GO:2000978 | negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.8 | 3.4 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.7 | 4.7 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.6 | 4.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.6 | 1.7 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.4 | 1.1 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of tolerance induction to self antigen(GO:0002649) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.3 | 1.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.3 | 1.0 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.3 | 6.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 1.1 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.3 | 2.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 | 1.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 | 0.6 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.2 | 1.1 | GO:0061181 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 2.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 0.7 | GO:0006507 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 | 0.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.8 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 6.6 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.1 | 0.4 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.1 | 0.5 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 1.6 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.3 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.7 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.8 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.7 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 1.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 2.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 3.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 1.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.0 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 2.6 | GO:1903831 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 2.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 3.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.7 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.6 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) guanylate kinase-associated protein clustering(GO:0097117) |
| 0.1 | 1.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.0 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 1.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 2.3 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 2.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 2.1 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.6 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.4 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 1.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 1.4 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 0.3 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 1.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 1.8 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 2.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.3 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 1.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.7 | GO:0071420 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 1.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.4 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.2 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 6.0 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.6 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 1.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 2.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.6 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.7 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 1.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.8 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 2.3 | GO:0016573 | histone acetylation(GO:0016573) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 6.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.5 | 4.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.4 | 1.9 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.3 | 1.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 1.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.2 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.2 | 2.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 2.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 1.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 4.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 1.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 1.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 6.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 7.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 2.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 3.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.1 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 4.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0071820 | N-box binding(GO:0071820) |
| 0.6 | 8.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.6 | 3.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.5 | 5.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 4.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 1.8 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.4 | 1.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.3 | 1.0 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.3 | 1.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 2.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 6.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 2.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.3 | 3.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 6.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 1.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 0.6 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.2 | 2.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 1.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 1.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 4.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 8.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 0.6 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 2.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 2.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 6.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 6.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 1.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.4 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 1.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.7 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.6 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.9 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 1.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 2.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 2.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 2.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 1.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 4.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 3.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 5.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 6.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 8.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 5.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 3.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 8.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 3.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 5.9 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.2 | 7.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 6.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 5.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 2.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |