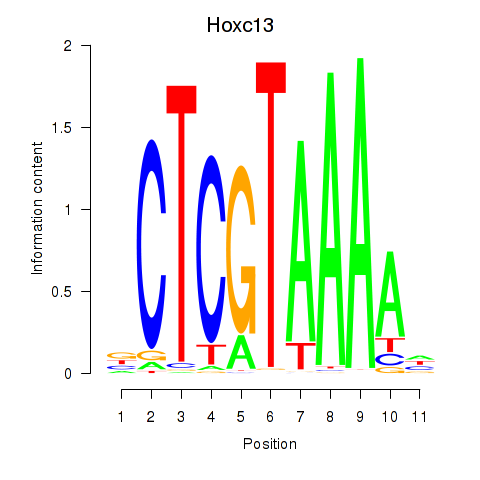
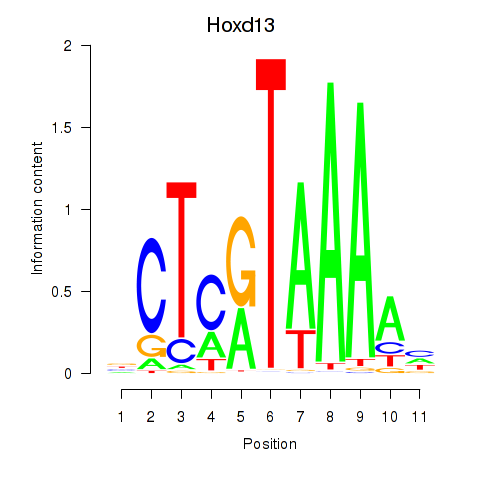
|
chr1_-_45542442
Show fit
|
8.78 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr9_+_92424276
Show fit
|
7.06 |
ENSMUST00000070522.14
ENSMUST00000160359.2
|
Plod2
|
procollagen lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr4_+_134042423
Show fit
|
6.18 |
ENSMUST00000105875.8
ENSMUST00000030638.7
|
Trim63
|
tripartite motif-containing 63
|
|
chr15_-_101759212
Show fit
|
5.88 |
ENSMUST00000023790.5
|
Krt1
|
keratin 1
|
|
chr6_-_52194440
Show fit
|
5.74 |
ENSMUST00000048715.9
|
Hoxa7
|
homeobox A7
|
|
chr4_-_133329479
Show fit
|
5.60 |
ENSMUST00000057311.4
|
Sfn
|
stratifin
|
|
chr17_+_75485906
Show fit
|
5.16 |
ENSMUST00000112514.2
|
Ltbp1
|
latent transforming growth factor beta binding protein 1
|
|
chr16_+_34815177
Show fit
|
5.04 |
ENSMUST00000231589.2
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr6_+_24528143
Show fit
|
4.82 |
ENSMUST00000031696.10
|
Asb15
|
ankyrin repeat and SOCS box-containing 15
|
|
chr11_+_96177449
Show fit
|
4.77 |
ENSMUST00000049352.8
|
Hoxb7
|
homeobox B7
|
|
chr17_+_75485791
Show fit
|
4.65 |
ENSMUST00000135447.8
ENSMUST00000112516.8
|
Ltbp1
|
latent transforming growth factor beta binding protein 1
|
|
chr6_-_52195663
Show fit
|
4.19 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7
|
|
chr3_+_79793237
Show fit
|
4.16 |
ENSMUST00000029567.9
|
Gask1b
|
golgi associated kinase 1B
|
|
chr11_+_96214078
Show fit
|
4.00 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3
|
|
chr2_+_74528071
Show fit
|
3.98 |
ENSMUST00000059272.10
|
Hoxd9
|
homeobox D9
|
|
chr14_+_65903840
Show fit
|
3.97 |
ENSMUST00000022610.15
|
Scara5
|
scavenger receptor class A, member 5
|
|
chr8_+_15107646
Show fit
|
3.94 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2
|
|
chr10_+_70040483
Show fit
|
3.74 |
ENSMUST00000020090.8
|
Mrln
|
myoregulin
|
|
chr7_-_30312246
Show fit
|
3.69 |
ENSMUST00000006476.6
|
Upk1a
|
uroplakin 1A
|
|
chr10_+_56253418
Show fit
|
3.45 |
ENSMUST00000068581.9
ENSMUST00000217789.2
|
Gja1
|
gap junction protein, alpha 1
|
|
chr10_+_28544356
Show fit
|
3.37 |
ENSMUST00000060409.13
ENSMUST00000056097.11
ENSMUST00000105516.9
|
Themis
|
thymocyte selection associated
|
|
chr5_+_17779273
Show fit
|
2.91 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr2_-_17465410
Show fit
|
2.89 |
ENSMUST00000145492.2
|
Nebl
|
nebulette
|
|
chr17_-_31363245
Show fit
|
2.86 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1)
|
|
chr12_+_75355082
Show fit
|
2.64 |
ENSMUST00000118602.8
ENSMUST00000118966.8
ENSMUST00000055390.6
|
Rhoj
|
ras homolog family member J
|
|
chr6_-_52194414
Show fit
|
2.54 |
ENSMUST00000140316.2
|
Hoxa7
|
homeobox A7
|
|
chr10_+_70040504
Show fit
|
2.48 |
ENSMUST00000190199.2
|
Mrln
|
myoregulin
|
|
chr14_+_65903878
Show fit
|
2.46 |
ENSMUST00000069226.7
|
Scara5
|
scavenger receptor class A, member 5
|
|
chr4_-_141553306
Show fit
|
2.42 |
ENSMUST00000102481.4
|
Cela2a
|
chymotrypsin-like elastase family, member 2A
|
|
chr15_-_101833160
Show fit
|
2.41 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4
|
|
chr1_-_74163575
Show fit
|
2.39 |
ENSMUST00000169786.8
ENSMUST00000212888.2
ENSMUST00000191104.7
|
Tns1
|
tensin 1
|
|
chr5_-_122639840
Show fit
|
2.28 |
ENSMUST00000177974.8
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr5_+_114284585
Show fit
|
2.25 |
ENSMUST00000102582.8
|
Acacb
|
acetyl-Coenzyme A carboxylase beta
|
|
chr11_+_96085118
Show fit
|
2.21 |
ENSMUST00000062709.4
|
Hoxb13
|
homeobox B13
|
|
chr3_+_126391046
Show fit
|
2.13 |
ENSMUST00000106401.8
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta
|
|
chr7_+_28240262
Show fit
|
2.01 |
ENSMUST00000119180.4
|
Sycn
|
syncollin
|
|
chr4_-_148584906
Show fit
|
2.01 |
ENSMUST00000030840.4
|
Angptl7
|
angiopoietin-like 7
|
|
chr1_-_156766351
Show fit
|
1.99 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr5_+_128677863
Show fit
|
1.93 |
ENSMUST00000117102.4
|
Fzd10
|
frizzled class receptor 10
|
|
chr1_-_39844467
Show fit
|
1.90 |
ENSMUST00000171319.4
|
Gm3646
|
predicted gene 3646
|
|
chr3_+_126390951
Show fit
|
1.88 |
ENSMUST00000171289.8
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta
|
|
chr15_-_101441254
Show fit
|
1.85 |
ENSMUST00000023720.8
|
Krt84
|
keratin 84
|
|
chr2_+_35022178
Show fit
|
1.75 |
ENSMUST00000113034.8
ENSMUST00000113037.10
ENSMUST00000150807.6
ENSMUST00000113033.6
|
Cntrl
|
centriolin
|
|
chr11_-_43492367
Show fit
|
1.64 |
ENSMUST00000020672.5
|
Fabp6
|
fatty acid binding protein 6
|
|
chr17_-_52133594
Show fit
|
1.63 |
ENSMUST00000129667.8
ENSMUST00000169480.8
ENSMUST00000148559.2
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr1_+_60948149
Show fit
|
1.56 |
ENSMUST00000027164.9
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4
|
|
chr1_+_107289659
Show fit
|
1.55 |
ENSMUST00000027566.3
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11
|
|
chr15_-_101482320
Show fit
|
1.55 |
ENSMUST00000042957.6
|
Krt75
|
keratin 75
|
|
chr9_+_32607301
Show fit
|
1.46 |
ENSMUST00000034534.13
ENSMUST00000050797.14
ENSMUST00000184887.2
|
Ets1
|
E26 avian leukemia oncogene 1, 5' domain
|
|
chr2_+_138120401
Show fit
|
1.43 |
ENSMUST00000075410.5
|
Btbd3
|
BTB (POZ) domain containing 3
|
|
chr13_-_19917092
Show fit
|
1.39 |
ENSMUST00000151029.3
|
Gpr141b
|
G protein-coupled receptor 141B
|
|
chr1_+_72346572
Show fit
|
1.32 |
ENSMUST00000027379.10
|
Xrcc5
|
X-ray repair complementing defective repair in Chinese hamster cells 5
|
|
chr11_-_99996452
Show fit
|
1.30 |
ENSMUST00000107416.3
|
Krt36
|
keratin 36
|
|
chr19_-_7319157
Show fit
|
1.30 |
ENSMUST00000164205.8
ENSMUST00000165286.8
ENSMUST00000168324.2
ENSMUST00000032557.15
|
Mark2
|
MAP/microtubule affinity regulating kinase 2
|
|
chr6_+_83063348
Show fit
|
1.30 |
ENSMUST00000041265.4
|
Lbx2
|
ladybird homeobox 2
|
|
chr14_-_103584179
Show fit
|
1.29 |
ENSMUST00000159855.8
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase
|
|
chr8_-_70591746
Show fit
|
1.25 |
ENSMUST00000212320.2
|
Rfxank
|
regulatory factor X-associated ankyrin-containing protein
|
|
chr9_-_77255069
Show fit
|
1.16 |
ENSMUST00000184848.8
ENSMUST00000184415.8
|
Mlip
|
muscular LMNA-interacting protein
|
|
chr6_-_24528012
Show fit
|
1.14 |
ENSMUST00000023851.9
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5
|
|
chr9_-_77255099
Show fit
|
1.12 |
ENSMUST00000184138.8
ENSMUST00000184006.8
ENSMUST00000185144.8
ENSMUST00000034910.16
|
Mlip
|
muscular LMNA-interacting protein
|
|
chr10_+_28544556
Show fit
|
1.09 |
ENSMUST00000161345.2
|
Themis
|
thymocyte selection associated
|
|
chr15_+_78867241
Show fit
|
1.08 |
ENSMUST00000109687.9
ENSMUST00000130663.10
|
Triobp
|
TRIO and F-actin binding protein
|
|
chrX_+_162923474
Show fit
|
1.03 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2
|
|
chr1_-_156766381
Show fit
|
1.03 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr1_-_171854818
Show fit
|
1.02 |
ENSMUST00000138714.2
ENSMUST00000027837.13
ENSMUST00000111264.8
|
Vangl2
|
VANGL planar cell polarity 2
|
|
chr17_+_46961250
Show fit
|
1.01 |
ENSMUST00000043464.14
|
Cul7
|
cullin 7
|
|
chrX_+_95498965
Show fit
|
0.98 |
ENSMUST00000033553.14
|
Heph
|
hephaestin
|
|
chr2_-_73316053
Show fit
|
0.96 |
ENSMUST00000102680.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1
|
|
chr13_-_40887444
Show fit
|
0.93 |
ENSMUST00000224665.2
|
Tfap2a
|
transcription factor AP-2, alpha
|
|
chr5_+_92957231
Show fit
|
0.92 |
ENSMUST00000113054.9
|
Shroom3
|
shroom family member 3
|
|
chr10_+_56253531
Show fit
|
0.92 |
ENSMUST00000220194.2
ENSMUST00000218834.2
|
Gja1
|
gap junction protein, alpha 1
|
|
chr4_-_136329953
Show fit
|
0.92 |
ENSMUST00000105847.8
ENSMUST00000116273.9
|
Kdm1a
|
lysine (K)-specific demethylase 1A
|
|
chr15_+_98972850
Show fit
|
0.89 |
ENSMUST00000039665.8
|
Troap
|
trophinin associated protein
|
|
chr4_-_63090355
Show fit
|
0.86 |
ENSMUST00000156618.9
ENSMUST00000030042.3
|
Kif12
|
kinesin family member 12
|
|
chr11_-_88608958
Show fit
|
0.86 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2
|
|
chr5_-_86893645
Show fit
|
0.85 |
ENSMUST00000161306.2
|
Tmprss11e
|
transmembrane protease, serine 11e
|
|
chr7_-_102408573
Show fit
|
0.83 |
ENSMUST00000210453.3
ENSMUST00000232246.3
ENSMUST00000239110.2
ENSMUST00000060187.15
ENSMUST00000168007.3
|
Olfr560
Olfr78
|
olfactory receptor 560
olfactory receptor 78
|
|
chr6_+_71886030
Show fit
|
0.83 |
ENSMUST00000055296.11
|
Polr1a
|
polymerase (RNA) I polypeptide A
|
|
chr15_-_101459092
Show fit
|
0.81 |
ENSMUST00000023713.10
|
Krt82
|
keratin 82
|
|
chr9_+_5308828
Show fit
|
0.79 |
ENSMUST00000162846.8
ENSMUST00000027012.14
|
Casp4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chr3_-_88317601
Show fit
|
0.78 |
ENSMUST00000193338.6
ENSMUST00000056370.13
|
Pmf1
|
polyamine-modulated factor 1
|
|
chr14_+_32043944
Show fit
|
0.77 |
ENSMUST00000022480.8
ENSMUST00000228529.2
|
Ogdhl
|
oxoglutarate dehydrogenase-like
|
|
chr9_+_110948492
Show fit
|
0.73 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2
|
|
chr11_+_77107006
Show fit
|
0.70 |
ENSMUST00000156488.8
ENSMUST00000037912.12
|
Ssh2
|
slingshot protein phosphatase 2
|
|
chr10_-_35587888
Show fit
|
0.65 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2
|
|
chr15_+_6552270
Show fit
|
0.64 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein
|
|
chr9_-_77255171
Show fit
|
0.63 |
ENSMUST00000185039.8
|
Mlip
|
muscular LMNA-interacting protein
|
|
chr15_-_101336669
Show fit
|
0.60 |
ENSMUST00000081945.5
|
Krt87
|
keratin 87
|
|
chr9_-_50571080
Show fit
|
0.60 |
ENSMUST00000034567.4
|
Dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex)
|
|
chr1_+_60948307
Show fit
|
0.58 |
ENSMUST00000097720.4
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4
|
|
chr7_+_67297152
Show fit
|
0.57 |
ENSMUST00000032774.16
ENSMUST00000107471.8
|
Ttc23
|
tetratricopeptide repeat domain 23
|
|
chr15_-_78947038
Show fit
|
0.56 |
ENSMUST00000151889.8
ENSMUST00000040676.11
|
Ankrd54
|
ankyrin repeat domain 54
|
|
chr10_+_127851031
Show fit
|
0.55 |
ENSMUST00000178041.8
ENSMUST00000026461.8
|
Prim1
|
DNA primase, p49 subunit
|
|
chr4_-_82768958
Show fit
|
0.54 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21
|
|
chr1_+_85538554
Show fit
|
0.52 |
ENSMUST00000162925.2
|
Sp140
|
Sp140 nuclear body protein
|
|
chr11_-_100510992
Show fit
|
0.49 |
ENSMUST00000014339.15
ENSMUST00000239490.2
ENSMUST00000239410.2
|
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7
|
|
chr2_+_112096154
Show fit
|
0.47 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chr3_-_49711706
Show fit
|
0.47 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18
|
|
chr11_-_99877423
Show fit
|
0.47 |
ENSMUST00000105050.4
|
Krtap16-1
|
keratin associated protein 16-1
|
|
chr17_+_37274714
Show fit
|
0.44 |
ENSMUST00000209623.2
|
Polr1has
|
RNA polymerase I subunit H, antisense
|
|
chr10_-_107321938
Show fit
|
0.43 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5
|
|
chr1_+_53352780
Show fit
|
0.43 |
ENSMUST00000027265.10
ENSMUST00000114484.8
|
Osgepl1
|
O-sialoglycoprotein endopeptidase-like 1
|
|
chr17_-_36149100
Show fit
|
0.41 |
ENSMUST00000134978.3
|
Tubb5
|
tubulin, beta 5 class I
|
|
chr13_+_35843816
Show fit
|
0.40 |
ENSMUST00000075220.14
|
Cdyl
|
chromodomain protein, Y chromosome-like
|
|
chr2_-_127634387
Show fit
|
0.40 |
ENSMUST00000135091.2
|
Mtln
|
mitoregulin
|
|
chr8_-_70591799
Show fit
|
0.39 |
ENSMUST00000075724.9
|
Rfxank
|
regulatory factor X-associated ankyrin-containing protein
|
|
chrX_-_7185424
Show fit
|
0.38 |
ENSMUST00000115746.8
|
Clcn5
|
chloride channel, voltage-sensitive 5
|
|
chr19_-_7319211
Show fit
|
0.38 |
ENSMUST00000171393.8
|
Mark2
|
MAP/microtubule affinity regulating kinase 2
|
|
chr18_-_46413886
Show fit
|
0.36 |
ENSMUST00000236999.2
|
Pggt1b
|
protein geranylgeranyltransferase type I, beta subunit
|
|
chr13_-_21586858
Show fit
|
0.32 |
ENSMUST00000117721.8
ENSMUST00000070785.16
ENSMUST00000116433.2
ENSMUST00000223831.2
ENSMUST00000116434.11
ENSMUST00000224820.2
|
Zkscan3
|
zinc finger with KRAB and SCAN domains 3
|
|
chr8_-_25528972
Show fit
|
0.32 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4
|
|
chrX_-_7185529
Show fit
|
0.31 |
ENSMUST00000128319.2
|
Clcn5
|
chloride channel, voltage-sensitive 5
|
|
chr17_+_35658131
Show fit
|
0.31 |
ENSMUST00000071951.14
ENSMUST00000116598.10
ENSMUST00000078205.14
ENSMUST00000076256.8
|
H2-Q7
|
histocompatibility 2, Q region locus 7
|
|
chr10_-_8394546
Show fit
|
0.30 |
ENSMUST00000061601.9
|
Ust
|
uronyl-2-sulfotransferase
|
|
chr9_-_63929308
Show fit
|
0.28 |
ENSMUST00000179458.2
ENSMUST00000041029.6
|
Smad6
|
SMAD family member 6
|
|
chr1_-_156766957
Show fit
|
0.28 |
ENSMUST00000171292.8
ENSMUST00000063199.13
ENSMUST00000027886.14
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr11_+_60066681
Show fit
|
0.27 |
ENSMUST00000102688.2
|
Rai1
|
retinoic acid induced 1
|
|
chr7_-_28330322
Show fit
|
0.27 |
ENSMUST00000040112.5
ENSMUST00000239470.2
|
Acp7
|
acid phosphatase 7, tartrate resistant
|
|
chr4_+_21879660
Show fit
|
0.26 |
ENSMUST00000029909.3
|
Coq3
|
coenzyme Q3 methyltransferase
|
|
chr15_+_66542598
Show fit
|
0.23 |
ENSMUST00000065916.14
|
Tg
|
thyroglobulin
|
|
chr1_+_83022653
Show fit
|
0.23 |
ENSMUST00000222567.2
|
Gm47969
|
predicted gene, 47969
|
|
chr3_+_88528410
Show fit
|
0.22 |
ENSMUST00000029694.14
ENSMUST00000176804.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2
|
|
chrX_+_41241049
Show fit
|
0.22 |
ENSMUST00000128799.3
|
Stag2
|
stromal antigen 2
|
|
chr8_+_96058907
Show fit
|
0.22 |
ENSMUST00000034245.16
|
Usb1
|
U6 snRNA biogenesis 1
|
|
chr18_+_44249254
Show fit
|
0.17 |
ENSMUST00000212114.2
|
Gm37797
|
predicted gene, 37797
|
|
chr2_+_20727274
Show fit
|
0.15 |
ENSMUST00000114607.8
|
Etl4
|
enhancer trap locus 4
|
|
chr7_-_28329924
Show fit
|
0.15 |
ENSMUST00000159095.2
ENSMUST00000159418.8
ENSMUST00000159560.3
|
Acp7
|
acid phosphatase 7, tartrate resistant
|
|
chr18_+_69478893
Show fit
|
0.15 |
ENSMUST00000202354.4
|
Tcf4
|
transcription factor 4
|
|
chr8_+_105991280
Show fit
|
0.14 |
ENSMUST00000036221.12
|
Fbxl8
|
F-box and leucine-rich repeat protein 8
|
|
chr11_-_23845207
Show fit
|
0.11 |
ENSMUST00000102863.3
ENSMUST00000020513.10
|
Papolg
|
poly(A) polymerase gamma
|
|
chr17_-_36149142
Show fit
|
0.11 |
ENSMUST00000001566.10
|
Tubb5
|
tubulin, beta 5 class I
|
|
chr3_+_102642516
Show fit
|
0.10 |
ENSMUST00000119902.6
|
Tspan2
|
tetraspanin 2
|
|
chr15_-_79658608
Show fit
|
0.10 |
ENSMUST00000229644.2
ENSMUST00000023055.8
|
Dnal4
|
dynein, axonemal, light chain 4
|
|
chr9_-_96601574
Show fit
|
0.10 |
ENSMUST00000128269.8
|
Zbtb38
|
zinc finger and BTB domain containing 38
|
|
chr15_-_79658584
Show fit
|
0.09 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4
|
|
chr9_+_38119661
Show fit
|
0.09 |
ENSMUST00000211975.3
|
Olfr893
|
olfactory receptor 893
|
|
chr3_-_75359125
Show fit
|
0.08 |
ENSMUST00000204341.3
|
Wdr49
|
WD repeat domain 49
|
|
chrX_+_7750558
Show fit
|
0.08 |
ENSMUST00000208640.2
ENSMUST00000207114.2
ENSMUST00000208633.2
ENSMUST00000208397.2
ENSMUST00000153620.3
ENSMUST00000123277.8
|
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2
|
|
chr4_-_44710408
Show fit
|
0.07 |
ENSMUST00000134968.9
ENSMUST00000173821.8
ENSMUST00000174319.8
ENSMUST00000173733.8
ENSMUST00000172866.8
ENSMUST00000165417.9
ENSMUST00000107825.9
ENSMUST00000102932.10
ENSMUST00000107827.9
ENSMUST00000107826.9
ENSMUST00000014174.14
|
Pax5
|
paired box 5
|
|
chr11_-_99433984
Show fit
|
0.07 |
ENSMUST00000107443.8
ENSMUST00000074253.4
|
Krt40
|
keratin 40
|
|
chr18_+_36877709
Show fit
|
0.04 |
ENSMUST00000007042.6
ENSMUST00000237095.2
|
Ik
|
IK cytokine
|
|
chr8_-_37200051
Show fit
|
0.02 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1
|
|
chr7_-_130748035
Show fit
|
0.02 |
ENSMUST00000070980.4
|
4933402N03Rik
|
RIKEN cDNA 4933402N03 gene
|
|
chr9_-_110775143
Show fit
|
0.01 |
ENSMUST00000199782.2
ENSMUST00000035075.13
|
Tdgf1
|
teratocarcinoma-derived growth factor 1
|
|
chr5_+_86952072
Show fit
|
0.01 |
ENSMUST00000119339.8
ENSMUST00000120498.8
|
Ythdc1
|
YTH domain containing 1
|
|
chr18_-_84854841
Show fit
|
0.00 |
ENSMUST00000236689.2
|
Gm17266
|
predicted gene, 17266
|
|
chr5_+_150119860
Show fit
|
0.00 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein
|
|
chr16_-_57051727
Show fit
|
0.00 |
ENSMUST00000226586.2
|
Tbc1d23
|
TBC1 domain family, member 23
|