Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
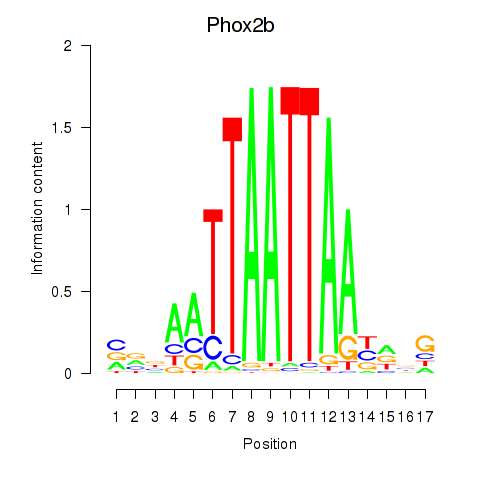
Results for Hoxc4_Arx_Otp_Esx1_Phox2b
Z-value: 0.47
Transcription factors associated with Hoxc4_Arx_Otp_Esx1_Phox2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
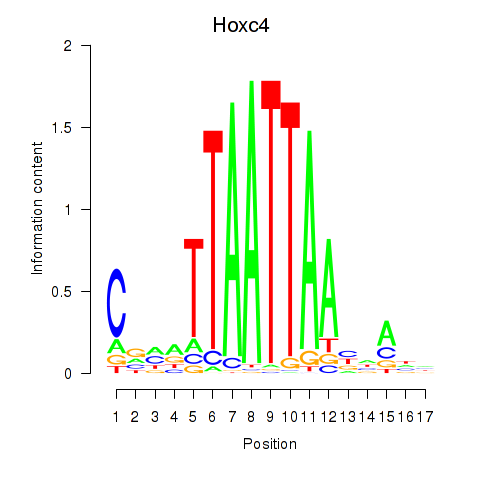
Hoxc4
|
ENSMUSG00000075394.5 | Hoxc4 |
|
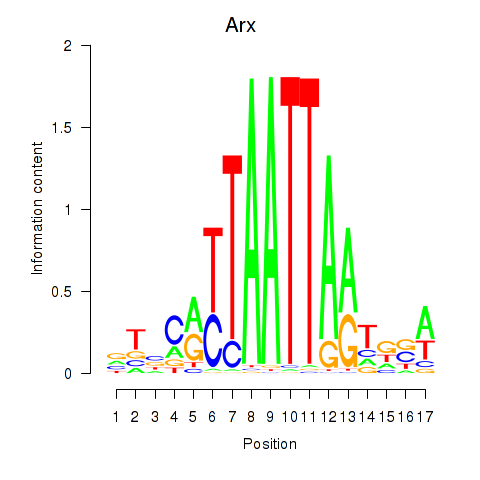
Arx
|
ENSMUSG00000035277.16 | Arx |
|
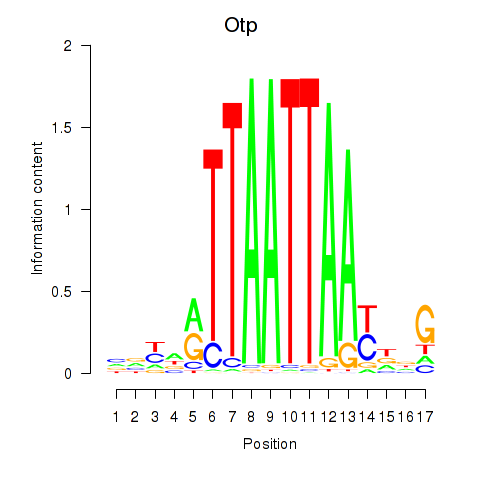
Otp
|
ENSMUSG00000021685.13 | Otp |
|
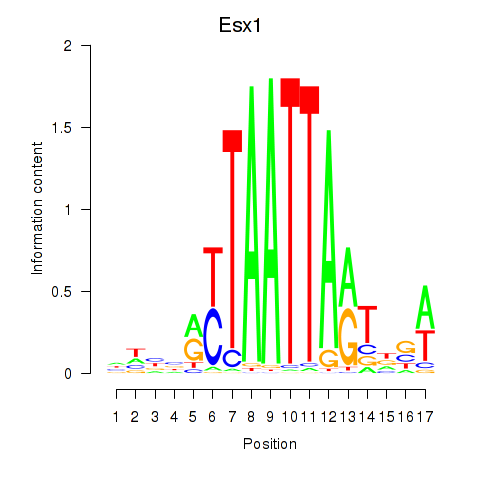
Esx1
|
ENSMUSG00000023443.14 | Esx1 |
|
Phox2b
|
ENSMUSG00000012520.11 | Phox2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Phox2b | mm39_v1_chr5_-_67256578_67256644 | -0.38 | 9.4e-04 | Click! |
| Esx1 | mm39_v1_chrX_-_136020873_136020920 | -0.13 | 2.6e-01 | Click! |
| Otp | mm39_v1_chr13_+_95012107_95012119 | -0.09 | 4.4e-01 | Click! |
| Arx | mm39_v1_chrX_+_92330102_92330128 | -0.04 | 7.5e-01 | Click! |
| Hoxc4 | mm39_v1_chr15_+_102927366_102927405 | -0.04 | 7.7e-01 | Click! |
Activity profile of Hoxc4_Arx_Otp_Esx1_Phox2b motif
Sorted Z-values of Hoxc4_Arx_Otp_Esx1_Phox2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc4_Arx_Otp_Esx1_Phox2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_39801188 | 5.46 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr19_-_39637489 | 4.49 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr19_-_39875192 | 4.47 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr15_-_82678490 | 3.22 |
ENSMUST00000006094.6
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26 |
| chr3_-_67422821 | 3.06 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr10_+_32959472 | 2.55 |
ENSMUST00000095762.5
ENSMUST00000218281.2 ENSMUST00000217779.2 ENSMUST00000219665.2 ENSMUST00000219931.2 |
Trdn
|
triadin |
| chr2_-_34990689 | 2.08 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chrM_+_10167 | 1.74 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr1_-_72323407 | 1.71 |
ENSMUST00000097698.5
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr1_-_72323464 | 1.65 |
ENSMUST00000027381.13
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr17_+_79919267 | 1.59 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chrM_+_9870 | 1.58 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr2_-_73410632 | 1.55 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chrX_-_158921370 | 1.51 |
ENSMUST00000033662.9
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chr9_+_21634779 | 1.51 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr3_+_7494108 | 1.43 |
ENSMUST00000193330.2
|
Pkia
|
protein kinase inhibitor, alpha |
| chr14_+_55797468 | 1.41 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr8_+_46463633 | 1.41 |
ENSMUST00000110381.9
|
Lrp2bp
|
Lrp2 binding protein |
| chr11_+_58062467 | 1.40 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr10_-_53252210 | 1.39 |
ENSMUST00000095691.7
|
Cep85l
|
centrosomal protein 85-like |
| chr9_-_70842090 | 1.37 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr7_-_44753168 | 1.31 |
ENSMUST00000211085.2
ENSMUST00000210642.2 ENSMUST00000003512.9 |
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr11_-_107228382 | 1.30 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr14_+_26616514 | 1.24 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr18_+_31742565 | 1.20 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr18_-_10706701 | 1.15 |
ENSMUST00000002549.9
ENSMUST00000117726.9 ENSMUST00000117828.9 |
Abhd3
|
abhydrolase domain containing 3 |
| chr14_+_55797443 | 1.15 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr10_+_97318223 | 1.12 |
ENSMUST00000163448.4
|
Dcn
|
decorin |
| chr7_-_12829100 | 1.12 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr5_+_90708962 | 1.12 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr7_-_126275529 | 1.09 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr4_+_108576846 | 1.01 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr14_+_40826970 | 0.97 |
ENSMUST00000225720.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr1_+_87983099 | 0.93 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr2_+_69500444 | 0.91 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr4_-_14621669 | 0.91 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr7_+_26819334 | 0.89 |
ENSMUST00000003100.10
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr7_+_37882642 | 0.88 |
ENSMUST00000178207.10
ENSMUST00000179525.10 |
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr3_-_72875187 | 0.88 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr6_-_41752111 | 0.87 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr19_-_45224251 | 0.86 |
ENSMUST00000099401.6
|
Lbx1
|
ladybird homeobox 1 |
| chr4_+_98919183 | 0.86 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr7_+_37883074 | 0.85 |
ENSMUST00000178876.10
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr2_+_69727599 | 0.80 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr14_-_30665232 | 0.80 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr15_-_66985760 | 0.80 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr7_-_14180496 | 0.79 |
ENSMUST00000063509.11
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr2_+_69727563 | 0.79 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr6_+_37847721 | 0.79 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr1_+_87983189 | 0.78 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr2_-_168608949 | 0.78 |
ENSMUST00000075044.10
|
Sall4
|
spalt like transcription factor 4 |
| chr3_+_66127330 | 0.78 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chr4_+_102446883 | 0.77 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr2_-_168609110 | 0.77 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr7_+_37883216 | 0.71 |
ENSMUST00000177983.2
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr16_+_22737227 | 0.69 |
ENSMUST00000231880.2
|
Fetub
|
fetuin beta |
| chr16_+_22737128 | 0.69 |
ENSMUST00000170805.9
|
Fetub
|
fetuin beta |
| chr1_-_63215952 | 0.67 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr2_-_111820618 | 0.67 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr9_-_55419442 | 0.66 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr14_-_66361931 | 0.66 |
ENSMUST00000070515.2
|
Ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr16_+_22737050 | 0.66 |
ENSMUST00000231768.2
|
Fetub
|
fetuin beta |
| chr1_-_63215812 | 0.65 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr19_+_11674413 | 0.64 |
ENSMUST00000069760.13
ENSMUST00000119053.3 |
Oosp3
|
oocyte secreted protein 3 |
| chr10_+_62860094 | 0.63 |
ENSMUST00000124784.8
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr5_-_3697806 | 0.63 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr4_+_43493344 | 0.63 |
ENSMUST00000030181.12
ENSMUST00000107922.3 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr11_+_73489420 | 0.63 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chr4_-_14621497 | 0.62 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr13_-_32967937 | 0.61 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chrM_+_14138 | 0.59 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chrX_+_100492684 | 0.58 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr7_+_17979272 | 0.58 |
ENSMUST00000066780.5
|
Mill1
|
MHC I like leukocyte 1 |
| chr16_-_19425453 | 0.58 |
ENSMUST00000078603.3
ENSMUST00000218837.2 |
Olfr170
|
olfactory receptor 170 |
| chr10_+_23770586 | 0.56 |
ENSMUST00000041416.8
|
Vnn1
|
vanin 1 |
| chr12_+_10440755 | 0.56 |
ENSMUST00000020947.7
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr6_-_141801897 | 0.55 |
ENSMUST00000165990.8
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr16_-_19443851 | 0.54 |
ENSMUST00000079891.4
|
Olfr171
|
olfactory receptor 171 |
| chr5_+_87148697 | 0.54 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr19_+_41921903 | 0.53 |
ENSMUST00000224258.2
ENSMUST00000026154.9 ENSMUST00000224896.2 |
Zdhhc16
|
zinc finger, DHHC domain containing 16 |
| chr10_-_129965752 | 0.53 |
ENSMUST00000215217.2
ENSMUST00000214192.2 |
Olfr824
|
olfactory receptor 824 |
| chr10_+_62860291 | 0.53 |
ENSMUST00000020262.5
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr11_+_59197746 | 0.50 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr4_+_145397238 | 0.50 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr4_+_147637714 | 0.49 |
ENSMUST00000139784.8
ENSMUST00000143885.8 ENSMUST00000081742.7 |
Zfp985
|
zinc finger protein 985 |
| chr6_-_115569504 | 0.48 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr4_+_115458172 | 0.48 |
ENSMUST00000084342.6
|
Cyp4a32
|
cytochrome P450, family 4, subfamily a, polypeptide 32 |
| chr2_+_126398048 | 0.47 |
ENSMUST00000141482.3
|
Slc27a2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chr11_+_60428788 | 0.47 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr10_-_81243475 | 0.47 |
ENSMUST00000140916.8
|
Nfic
|
nuclear factor I/C |
| chr10_-_88440996 | 0.46 |
ENSMUST00000121629.8
|
Mybpc1
|
myosin binding protein C, slow-type |
| chr13_+_120616163 | 0.46 |
ENSMUST00000179071.2
|
Gm20767
|
predicted gene, 20767 |
| chr15_+_97990431 | 0.46 |
ENSMUST00000229280.2
ENSMUST00000163507.8 ENSMUST00000230445.2 |
Pfkm
|
phosphofructokinase, muscle |
| chr11_+_116734104 | 0.46 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr13_-_32960379 | 0.46 |
ENSMUST00000230119.2
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr3_+_94280101 | 0.45 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr17_-_37404764 | 0.44 |
ENSMUST00000087144.5
|
Olfr91
|
olfactory receptor 91 |
| chr1_+_165288606 | 0.44 |
ENSMUST00000027853.6
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr1_-_150341911 | 0.43 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr5_-_28415361 | 0.43 |
ENSMUST00000141196.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr3_+_132335704 | 0.43 |
ENSMUST00000212594.2
|
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr6_-_57512355 | 0.43 |
ENSMUST00000042766.6
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chr4_+_95445731 | 0.41 |
ENSMUST00000079223.11
ENSMUST00000177394.8 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr14_+_69585036 | 0.39 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr8_-_62355690 | 0.39 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr2_+_152318969 | 0.39 |
ENSMUST00000109836.3
|
Defb20
|
defensin beta 20 |
| chr10_+_127226180 | 0.39 |
ENSMUST00000077046.12
ENSMUST00000105250.9 |
R3hdm2
|
R3H domain containing 2 |
| chr10_+_127734384 | 0.39 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr7_-_44752508 | 0.38 |
ENSMUST00000209830.2
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr11_-_100653754 | 0.38 |
ENSMUST00000107360.3
ENSMUST00000055083.4 |
Hcrt
|
hypocretin |
| chr14_-_45626237 | 0.38 |
ENSMUST00000227865.2
ENSMUST00000226856.2 ENSMUST00000226276.2 ENSMUST00000046191.9 |
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr15_+_98350469 | 0.37 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281 |
| chr1_-_163552693 | 0.37 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chrX_+_16485937 | 0.35 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr9_-_85631361 | 0.35 |
ENSMUST00000039213.15
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr17_+_29493049 | 0.34 |
ENSMUST00000149405.4
|
BC004004
|
cDNA sequence BC004004 |
| chr5_-_28415020 | 0.34 |
ENSMUST00000118882.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr14_-_63221950 | 0.33 |
ENSMUST00000100493.3
|
Defb48
|
defensin beta 48 |
| chr6_+_78347636 | 0.33 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr10_+_29074950 | 0.33 |
ENSMUST00000217011.2
|
Gm49353
|
predicted gene, 49353 |
| chr2_-_62242562 | 0.33 |
ENSMUST00000047812.8
|
Dpp4
|
dipeptidylpeptidase 4 |
| chr13_-_120374288 | 0.33 |
ENSMUST00000179502.2
|
Gm21761
|
predicted gene, 21761 |
| chr14_-_88708782 | 0.32 |
ENSMUST00000192557.2
ENSMUST00000061628.7 |
Pcdh20
|
protocadherin 20 |
| chr5_-_150441629 | 0.32 |
ENSMUST00000118769.2
|
Zar1l
|
zygote arrest 1-like |
| chr12_-_84664001 | 0.32 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chrX_+_102400061 | 0.32 |
ENSMUST00000116547.3
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr11_+_58311921 | 0.32 |
ENSMUST00000013797.3
|
1810065E05Rik
|
RIKEN cDNA 1810065E05 gene |
| chr7_-_46316767 | 0.32 |
ENSMUST00000168335.3
ENSMUST00000107669.9 |
Tph1
|
tryptophan hydroxylase 1 |
| chr18_+_4920513 | 0.31 |
ENSMUST00000126977.8
|
Svil
|
supervillin |
| chr1_-_174749379 | 0.31 |
ENSMUST00000055294.4
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chr7_-_101486983 | 0.31 |
ENSMUST00000185929.2
ENSMUST00000165052.8 |
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr11_-_49004584 | 0.31 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr2_+_20742115 | 0.30 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr17_+_37269468 | 0.30 |
ENSMUST00000040177.7
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr4_+_145241454 | 0.29 |
ENSMUST00000105741.2
|
Zfp990
|
zinc finger protein 990 |
| chr1_-_80191649 | 0.29 |
ENSMUST00000058748.2
|
Fam124b
|
family with sequence similarity 124, member B |
| chr5_+_31684331 | 0.29 |
ENSMUST00000114533.9
ENSMUST00000202214.4 ENSMUST00000201858.4 ENSMUST00000202950.4 |
Slc4a1ap
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr4_-_146993984 | 0.29 |
ENSMUST00000238583.2
ENSMUST00000049821.4 |
Gm21411
|
predicted gene, 21411 |
| chr2_+_89757653 | 0.29 |
ENSMUST00000213720.3
ENSMUST00000102609.3 |
Olfr1258
|
olfactory receptor 1258 |
| chr5_-_28415166 | 0.29 |
ENSMUST00000117098.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr7_+_45354512 | 0.29 |
ENSMUST00000080885.12
ENSMUST00000211513.2 ENSMUST00000211357.2 |
Dbp
|
D site albumin promoter binding protein |
| chr13_-_17869314 | 0.29 |
ENSMUST00000221598.2
ENSMUST00000068545.6 ENSMUST00000220514.2 |
Sugct
|
succinyl-CoA glutarate-CoA transferase |
| chr16_-_45664591 | 0.28 |
ENSMUST00000076333.12
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr1_+_173964312 | 0.28 |
ENSMUST00000053941.4
|
Olfr424
|
olfactory receptor 424 |
| chr7_-_103191924 | 0.28 |
ENSMUST00000214269.3
|
Olfr612
|
olfactory receptor 612 |
| chr11_+_50917831 | 0.27 |
ENSMUST00000072152.2
|
Olfr54
|
olfactory receptor 54 |
| chr7_-_10488291 | 0.27 |
ENSMUST00000226874.2
ENSMUST00000227003.2 ENSMUST00000228561.2 ENSMUST00000228248.2 ENSMUST00000228526.2 ENSMUST00000228098.2 ENSMUST00000227940.2 ENSMUST00000228374.2 ENSMUST00000227702.2 |
Vmn1r71
|
vomeronasal 1 receptor 71 |
| chr8_+_22329942 | 0.27 |
ENSMUST00000006745.4
|
Defb2
|
defensin beta 2 |
| chr9_-_79920131 | 0.27 |
ENSMUST00000217264.2
|
Filip1
|
filamin A interacting protein 1 |
| chr2_-_89678487 | 0.27 |
ENSMUST00000214428.3
|
Olfr48
|
olfactory receptor 48 |
| chr2_+_130037114 | 0.27 |
ENSMUST00000077988.8
|
Tmc2
|
transmembrane channel-like gene family 2 |
| chr2_-_71198091 | 0.27 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr17_+_29493157 | 0.26 |
ENSMUST00000234234.2
|
BC004004
|
cDNA sequence BC004004 |
| chr6_-_129077867 | 0.26 |
ENSMUST00000032258.8
|
Clec2e
|
C-type lectin domain family 2, member e |
| chr5_-_21087023 | 0.26 |
ENSMUST00000118174.8
|
Phtf2
|
putative homeodomain transcription factor 2 |
| chr18_+_4993795 | 0.26 |
ENSMUST00000153016.8
|
Svil
|
supervillin |
| chr4_+_22357543 | 0.25 |
ENSMUST00000039234.10
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr4_+_145595364 | 0.25 |
ENSMUST00000123460.2
|
Zfp986
|
zinc finger protein 986 |
| chr5_-_38637624 | 0.25 |
ENSMUST00000067886.12
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr3_+_121746862 | 0.25 |
ENSMUST00000037958.14
ENSMUST00000196904.5 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr9_-_78222078 | 0.24 |
ENSMUST00000113376.10
|
Omt2a
|
oocyte maturation, alpha |
| chr9_+_99333474 | 0.24 |
ENSMUST00000163199.5
|
Nme9
|
NME/NM23 family member 9 |
| chr1_+_88234454 | 0.24 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr9_+_38738911 | 0.24 |
ENSMUST00000051238.7
ENSMUST00000219798.2 |
Olfr923
|
olfactory receptor 923 |
| chr14_-_45626198 | 0.24 |
ENSMUST00000226590.2
|
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr6_+_145879839 | 0.24 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr6_-_30936013 | 0.24 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr17_+_29493113 | 0.23 |
ENSMUST00000234326.2
ENSMUST00000235117.2 |
BC004004
|
cDNA sequence BC004004 |
| chr7_+_30193047 | 0.23 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr17_+_37977879 | 0.23 |
ENSMUST00000215811.2
|
Olfr118
|
olfactory receptor 118 |
| chr19_-_29312046 | 0.22 |
ENSMUST00000044143.6
|
Rln1
|
relaxin 1 |
| chrX_-_74886623 | 0.22 |
ENSMUST00000114057.8
|
Pls3
|
plastin 3 (T-isoform) |
| chr7_-_5325456 | 0.22 |
ENSMUST00000207520.2
|
Nlrp2
|
NLR family, pyrin domain containing 2 |
| chr6_-_129752812 | 0.22 |
ENSMUST00000095409.3
|
Klrh1
|
killer cell lectin-like receptor subfamily H, member 1 |
| chr16_-_45664664 | 0.22 |
ENSMUST00000036355.13
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chrX_+_133587268 | 0.22 |
ENSMUST00000124226.3
|
Armcx4
|
armadillo repeat containing, X-linked 4 |
| chrM_-_14061 | 0.22 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr13_+_90237824 | 0.22 |
ENSMUST00000012566.9
|
Tmem167
|
transmembrane protein 167 |
| chr19_-_53933052 | 0.22 |
ENSMUST00000135402.4
|
Bbip1
|
BBSome interacting protein 1 |
| chr2_+_67935015 | 0.21 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr7_-_8164653 | 0.21 |
ENSMUST00000168807.3
|
Vmn2r41
|
vomeronasal 2, receptor 41 |
| chr12_-_118162652 | 0.21 |
ENSMUST00000084806.7
|
Dnah11
|
dynein, axonemal, heavy chain 11 |
| chr11_-_4045343 | 0.20 |
ENSMUST00000004868.6
|
Mtfp1
|
mitochondrial fission process 1 |
| chr17_-_37430949 | 0.20 |
ENSMUST00000214994.2
ENSMUST00000216341.2 |
Olfr92
|
olfactory receptor 92 |
| chrX_+_105059305 | 0.20 |
ENSMUST00000033582.5
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr7_-_102540089 | 0.20 |
ENSMUST00000217024.2
|
Olfr569
|
olfactory receptor 569 |
| chr8_-_49008305 | 0.20 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr16_-_19341016 | 0.20 |
ENSMUST00000214315.2
|
Olfr167
|
olfactory receptor 167 |
| chr2_-_86257093 | 0.20 |
ENSMUST00000217481.2
|
Olfr1062
|
olfactory receptor 1062 |
| chr4_+_147056433 | 0.20 |
ENSMUST00000146688.3
|
Zfp989
|
zinc finger protein 989 |
| chr8_-_22550035 | 0.19 |
ENSMUST00000110738.3
|
Atp7b
|
ATPase, Cu++ transporting, beta polypeptide |
| chr13_-_42001075 | 0.19 |
ENSMUST00000179758.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr10_-_20600797 | 0.19 |
ENSMUST00000020165.14
|
Pde7b
|
phosphodiesterase 7B |
| chr16_+_88525719 | 0.19 |
ENSMUST00000060494.8
|
Krtap13-1
|
keratin associated protein 13-1 |
| chr5_-_66672158 | 0.19 |
ENSMUST00000161879.8
ENSMUST00000159357.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr11_-_65160767 | 0.19 |
ENSMUST00000102635.10
|
Myocd
|
myocardin |
| chr10_-_23112973 | 0.19 |
ENSMUST00000218049.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr2_-_104847360 | 0.19 |
ENSMUST00000111110.3
ENSMUST00000028592.12 |
Eif3m
|
eukaryotic translation initiation factor 3, subunit M |
| chr4_+_146098479 | 0.19 |
ENSMUST00000131932.2
|
Zfp600
|
zinc finger protein 600 |
| chr12_+_55350023 | 0.19 |
ENSMUST00000184766.8
ENSMUST00000183475.8 ENSMUST00000183654.2 |
Prorp
|
protein only RNase P catalytic subunit |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.5 | 15.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.4 | 2.6 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.4 | 1.6 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.4 | 1.5 | GO:1905167 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.3 | 1.0 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 0.3 | 1.4 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.3 | 0.8 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.2 | 0.7 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.2 | 1.9 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.2 | 1.9 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.5 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.9 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 1.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 1.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:1900239 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.1 | 1.6 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.3 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.8 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.5 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.1 | 0.8 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 1.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.5 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.2 | GO:0060618 | nipple development(GO:0060618) |
| 0.1 | 0.6 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.2 | GO:0009107 | lipoate metabolic process(GO:0009106) lipoate biosynthetic process(GO:0009107) |
| 0.1 | 2.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 0.3 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 1.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 3.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 1.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 2.6 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.4 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.9 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.2 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.4 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 1.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.4 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.4 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.0 | 0.9 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 1.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.6 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.5 | GO:0031034 | myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.4 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.4 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.2 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.2 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.0 | 0.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.0 | GO:0044107 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.4 | GO:0097435 | fibril organization(GO:0097435) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.5 | 1.5 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.2 | 1.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 0.7 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 0.4 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 0.8 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 5.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 1.1 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 3.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 1.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.4 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.5 | 1.5 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.4 | 15.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.4 | 2.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 1.0 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 1.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 0.9 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 1.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 1.5 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.2 | 4.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 3.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.4 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.7 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.4 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 1.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.8 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.5 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.9 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 1.6 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 0.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.2 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 0.6 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.6 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.4 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.2 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.3 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.9 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.1 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 2.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |