Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
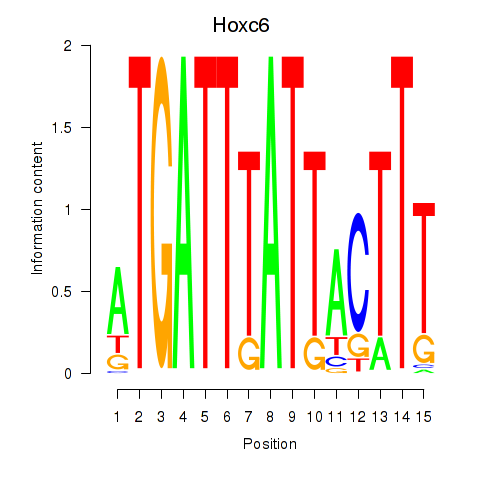
Results for Hoxc6
Z-value: 0.60
Transcription factors associated with Hoxc6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc6
|
ENSMUSG00000001661.6 | Hoxc6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc6 | mm39_v1_chr15_+_102917977_102918011 | 0.26 | 2.9e-02 | Click! |
Activity profile of Hoxc6 motif
Sorted Z-values of Hoxc6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_129518723 | 4.07 |
ENSMUST00000199615.5
ENSMUST00000197079.5 |
Egf
|
epidermal growth factor |
| chrX_+_162945162 | 2.91 |
ENSMUST00000131543.2
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr1_+_88062508 | 2.77 |
ENSMUST00000113134.8
ENSMUST00000140092.8 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr3_+_151143524 | 2.71 |
ENSMUST00000046977.12
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr3_+_151143557 | 2.61 |
ENSMUST00000196970.3
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr3_-_10273628 | 2.60 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr19_+_32597379 | 2.56 |
ENSMUST00000236290.2
ENSMUST00000025833.7 |
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr3_-_92393193 | 2.38 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr14_-_64654397 | 2.38 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr1_+_88030951 | 2.24 |
ENSMUST00000113135.6
ENSMUST00000113138.8 |
Ugt1a7c
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A7C UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr3_-_144738526 | 2.17 |
ENSMUST00000029919.7
|
Clca1
|
chloride channel accessory 1 |
| chrM_+_14138 | 1.95 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chrM_+_9459 | 1.92 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr1_+_19279138 | 1.89 |
ENSMUST00000027059.11
|
Tfap2b
|
transcription factor AP-2 beta |
| chr14_-_64654592 | 1.85 |
ENSMUST00000210363.2
|
Msra
|
methionine sulfoxide reductase A |
| chrM_-_14061 | 1.81 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr9_-_27066685 | 1.61 |
ENSMUST00000034472.16
|
Jam3
|
junction adhesion molecule 3 |
| chr1_+_19279178 | 1.60 |
ENSMUST00000187754.7
|
Tfap2b
|
transcription factor AP-2 beta |
| chr14_+_79086492 | 1.58 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr7_+_51537645 | 1.56 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr8_-_110688716 | 1.52 |
ENSMUST00000001722.14
ENSMUST00000051430.7 |
Marveld3
|
MARVEL (membrane-associating) domain containing 3 |
| chrM_+_8603 | 1.50 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr10_+_5543769 | 1.49 |
ENSMUST00000051809.10
|
Myct1
|
myc target 1 |
| chr19_+_24853039 | 1.43 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr7_+_45204317 | 1.41 |
ENSMUST00000107752.12
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr3_-_151455514 | 1.37 |
ENSMUST00000029671.9
|
Ifi44
|
interferon-induced protein 44 |
| chr2_+_128704867 | 1.35 |
ENSMUST00000110324.8
|
Fbln7
|
fibulin 7 |
| chr5_-_83502966 | 1.30 |
ENSMUST00000053543.11
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr6_+_36364990 | 1.29 |
ENSMUST00000172278.8
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr6_-_3494587 | 1.24 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chrX_-_93256244 | 1.19 |
ENSMUST00000113922.2
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr5_-_83502793 | 1.19 |
ENSMUST00000146669.2
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr18_-_56705960 | 1.15 |
ENSMUST00000174518.8
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr17_+_13094921 | 0.99 |
ENSMUST00000075296.4
|
Mrgprh
|
MAS-related GPR, member H |
| chr7_+_73390026 | 0.94 |
ENSMUST00000107456.4
|
Fam174b
|
family with sequence similarity 174, member B |
| chr2_-_174188505 | 0.92 |
ENSMUST00000168292.2
|
Gm20721
|
predicted gene, 20721 |
| chr1_-_189654535 | 0.92 |
ENSMUST00000027897.8
|
Smyd2
|
SET and MYND domain containing 2 |
| chr15_-_50752437 | 0.90 |
ENSMUST00000183997.8
ENSMUST00000183757.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chrX_-_93256291 | 0.88 |
ENSMUST00000050328.15
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr3_+_92840279 | 0.88 |
ENSMUST00000046234.5
|
Lce3b
|
late cornified envelope 3B |
| chr5_+_119970733 | 0.86 |
ENSMUST00000202723.4
|
Tbx5
|
T-box 5 |
| chr15_-_50753061 | 0.85 |
ENSMUST00000165201.9
ENSMUST00000184458.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr2_+_150628655 | 0.78 |
ENSMUST00000045441.8
|
Pygb
|
brain glycogen phosphorylase |
| chr11_-_70560110 | 0.74 |
ENSMUST00000129434.2
ENSMUST00000018431.13 |
Spag7
|
sperm associated antigen 7 |
| chr16_-_56533179 | 0.73 |
ENSMUST00000136394.8
|
Tfg
|
Trk-fused gene |
| chrX_-_135641869 | 0.72 |
ENSMUST00000166930.8
ENSMUST00000113095.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chrX_-_135642025 | 0.70 |
ENSMUST00000155207.8
ENSMUST00000080411.13 ENSMUST00000169418.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr10_-_129591354 | 0.67 |
ENSMUST00000059038.3
|
Olfr807
|
olfactory receptor 807 |
| chr14_+_75479727 | 0.66 |
ENSMUST00000022576.10
|
Cpb2
|
carboxypeptidase B2 (plasma) |
| chr7_+_45204350 | 0.65 |
ENSMUST00000210300.2
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr2_+_111329683 | 0.57 |
ENSMUST00000219064.3
|
Olfr1291-ps1
|
olfactory receptor 1291, pseudogene 1 |
| chr3_-_113325938 | 0.57 |
ENSMUST00000132353.2
|
Amy2a1
|
amylase 2a1 |
| chr15_+_44482667 | 0.53 |
ENSMUST00000228648.2
ENSMUST00000226165.2 |
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr4_+_33062999 | 0.52 |
ENSMUST00000108162.8
ENSMUST00000024035.9 |
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr10_-_62285458 | 0.52 |
ENSMUST00000020273.16
|
Supv3l1
|
suppressor of var1, 3-like 1 (S. cerevisiae) |
| chr2_-_89172273 | 0.51 |
ENSMUST00000215469.2
|
Olfr1233
|
olfactory receptor 1233 |
| chr19_+_12972378 | 0.50 |
ENSMUST00000207997.3
|
Olfr1451
|
olfactory receptor 1451 |
| chr2_-_111062182 | 0.48 |
ENSMUST00000099620.5
|
Olfr1275
|
olfactory receptor 1275 |
| chr11_+_73973733 | 0.47 |
ENSMUST00000178159.2
|
Zfp616
|
zinc finger protein 616 |
| chr5_-_121329385 | 0.46 |
ENSMUST00000054547.9
ENSMUST00000100770.9 |
Ptpn11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr17_+_38082190 | 0.45 |
ENSMUST00000217119.2
|
Olfr122
|
olfactory receptor 122 |
| chr2_-_51039112 | 0.44 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr15_+_91557378 | 0.40 |
ENSMUST00000060642.7
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr10_+_23814470 | 0.40 |
ENSMUST00000079134.5
|
Taar2
|
trace amine-associated receptor 2 |
| chr4_-_84464521 | 0.39 |
ENSMUST00000177040.2
|
Bnc2
|
basonuclin 2 |
| chr6_-_123830422 | 0.39 |
ENSMUST00000162046.3
|
Vmn2r25
|
vomeronasal 2, receptor 25 |
| chr2_-_87524291 | 0.38 |
ENSMUST00000077471.5
|
Olfr1136
|
olfactory receptor 1136 |
| chr8_-_84976330 | 0.38 |
ENSMUST00000019506.9
|
D8Ertd738e
|
DNA segment, Chr 8, ERATO Doi 738, expressed |
| chr8_+_46111778 | 0.36 |
ENSMUST00000143820.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_-_129894613 | 0.36 |
ENSMUST00000118060.8
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr11_-_94673526 | 0.35 |
ENSMUST00000100554.8
|
Tmem92
|
transmembrane protein 92 |
| chr7_-_13856967 | 0.34 |
ENSMUST00000098809.4
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr11_-_58624049 | 0.31 |
ENSMUST00000075607.5
|
Olfr317
|
olfactory receptor 317 |
| chr14_-_52593865 | 0.31 |
ENSMUST00000215147.2
|
Olfr1513
|
olfactory receptor 1513 |
| chr19_+_6414587 | 0.30 |
ENSMUST00000155973.2
|
Sf1
|
splicing factor 1 |
| chr6_-_132755125 | 0.29 |
ENSMUST00000065532.2
|
Tas2r136
|
taste receptor, type 2, member 136 |
| chr7_+_108064533 | 0.29 |
ENSMUST00000217616.3
|
Olfr498
|
olfactory receptor 498 |
| chr2_-_85193571 | 0.28 |
ENSMUST00000215511.2
|
Olfr988
|
olfactory receptor 988 |
| chr10_-_129463803 | 0.27 |
ENSMUST00000204979.3
|
Olfr798
|
olfactory receptor 798 |
| chrX_-_95400846 | 0.26 |
ENSMUST00000179832.8
|
Hsf3
|
heat shock transcription factor 3 |
| chr15_+_44482944 | 0.25 |
ENSMUST00000022964.9
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr10_+_19588318 | 0.24 |
ENSMUST00000020185.5
|
Il20ra
|
interleukin 20 receptor, alpha |
| chr2_-_121211410 | 0.24 |
ENSMUST00000038389.15
|
Strc
|
stereocilin |
| chr9_-_39237341 | 0.21 |
ENSMUST00000216132.2
|
Olfr948
|
olfactory receptor 948 |
| chr12_-_114226570 | 0.20 |
ENSMUST00000103479.4
ENSMUST00000195619.2 |
Ighv3-5
|
immunoglobulin heavy variable 3-5 |
| chrX_+_55958781 | 0.19 |
ENSMUST00000168724.2
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr9_-_76120939 | 0.19 |
ENSMUST00000074880.6
|
Gfral
|
GDNF family receptor alpha like |
| chr5_+_17779273 | 0.19 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr10_-_13199971 | 0.19 |
ENSMUST00000105543.9
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr3_+_133942244 | 0.15 |
ENSMUST00000181904.3
|
Cxxc4
|
CXXC finger 4 |
| chr19_+_11872971 | 0.14 |
ENSMUST00000072784.4
|
Olfr1420
|
olfactory receptor 1420 |
| chr14_-_52616625 | 0.13 |
ENSMUST00000214980.2
|
Olfr1512
|
olfactory receptor 1512 |
| chr14_+_26722319 | 0.13 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr2_-_88562710 | 0.12 |
ENSMUST00000213118.2
|
Olfr1197
|
olfactory receptor 1197 |
| chr17_+_3532455 | 0.09 |
ENSMUST00000227604.2
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr1_-_22386016 | 0.09 |
ENSMUST00000164877.8
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr9_+_19351562 | 0.09 |
ENSMUST00000217273.2
|
Olfr849
|
olfactory receptor 849 |
| chr9_-_39161290 | 0.09 |
ENSMUST00000213472.2
|
Olfr1537
|
olfactory receptor 1537 |
| chr6_-_57340712 | 0.08 |
ENSMUST00000228156.2
ENSMUST00000227186.2 ENSMUST00000228294.2 ENSMUST00000228342.2 |
Vmn1r17
|
vomeronasal 1 receptor 17 |
| chr3_+_145855929 | 0.08 |
ENSMUST00000098524.5
|
Mcoln2
|
mucolipin 2 |
| chr19_-_12870888 | 0.07 |
ENSMUST00000216805.3
ENSMUST00000207741.2 |
Olfr1446
|
olfactory receptor 1446 |
| chr5_-_129030367 | 0.05 |
ENSMUST00000111346.6
ENSMUST00000200470.5 |
Rimbp2
|
RIMS binding protein 2 |
| chr7_-_85974838 | 0.05 |
ENSMUST00000214977.2
|
Olfr308
|
olfactory receptor 308 |
| chr7_+_5023375 | 0.04 |
ENSMUST00000076251.7
|
Zfp865
|
zinc finger protein 865 |
| chrX_+_48552803 | 0.01 |
ENSMUST00000130558.8
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr2_-_89172742 | 0.00 |
ENSMUST00000216561.2
|
Olfr1233
|
olfactory receptor 1233 |
| chr2_-_88590813 | 0.00 |
ENSMUST00000124021.3
|
Olfr1199
|
olfactory receptor 1199 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0097273 | creatinine homeostasis(GO:0097273) cellular ammonia homeostasis(GO:0097275) cellular creatinine homeostasis(GO:0097276) cellular urea homeostasis(GO:0097277) |
| 0.7 | 2.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.7 | 2.9 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.6 | 2.6 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.5 | 4.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 0.9 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.3 | 2.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 1.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 1.9 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.2 | 0.5 | GO:2000850 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.2 | 0.7 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 2.6 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.4 | GO:2000469 | negative regulation of peroxidase activity(GO:2000469) |
| 0.1 | 0.5 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 1.6 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 2.5 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.5 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 2.1 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 2.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.8 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 1.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.0 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 1.9 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.8 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 2.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 1.5 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 4.2 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.4 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 2.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 1.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 1.9 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 0.4 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 2.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 3.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 4.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.6 | 2.6 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.5 | 2.9 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.4 | 2.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 1.3 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.3 | 2.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.3 | 1.2 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.2 | 1.9 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 0.8 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 4.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 3.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.0 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.1 | 5.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 3.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.4 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 2.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.3 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 1.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 2.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 2.5 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 2.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 4.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 2.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |