Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Hoxd1
Z-value: 1.03
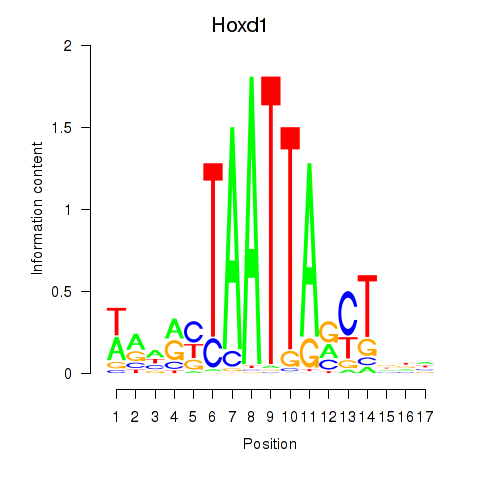
Transcription factors associated with Hoxd1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd1
|
ENSMUSG00000042448.6 | Hoxd1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd1 | mm39_v1_chr2_+_74593324_74593324 | 0.20 | 1.0e-01 | Click! |
Activity profile of Hoxd1 motif
Sorted Z-values of Hoxd1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_69282389 | 10.77 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr7_+_45271229 | 8.52 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr2_-_32277773 | 8.50 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr2_-_32278245 | 7.49 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr10_-_75946790 | 7.00 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr16_+_45044678 | 6.53 |
ENSMUST00000102802.10
ENSMUST00000063654.6 |
Btla
|
B and T lymphocyte associated |
| chr12_-_114487525 | 6.28 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr6_-_69584812 | 5.64 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr6_-_69415741 | 5.60 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr3_-_72875187 | 5.50 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr12_-_114443071 | 5.47 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr6_-_124756645 | 5.45 |
ENSMUST00000147669.2
ENSMUST00000128697.8 ENSMUST00000032218.10 ENSMUST00000112475.9 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr5_+_88127290 | 4.97 |
ENSMUST00000008051.5
|
Cabs1
|
calcium binding protein, spermatid specific 1 |
| chr7_-_103778992 | 4.92 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr7_-_126574959 | 4.87 |
ENSMUST00000206296.2
ENSMUST00000205437.3 |
Cdiptos
|
CDIP transferase, opposite strand |
| chr9_+_53678801 | 4.82 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr6_-_69355456 | 4.70 |
ENSMUST00000196595.2
|
Igkv4-63
|
immunoglobulin kappa variable 4-63 |
| chr3_-_15640045 | 4.47 |
ENSMUST00000192382.6
ENSMUST00000195778.3 ENSMUST00000091319.7 |
Sirpb1b
|
signal-regulatory protein beta 1B |
| chr6_-_69553484 | 4.41 |
ENSMUST00000103357.4
|
Igkv4-57
|
immunoglobulin kappa variable 4-57 |
| chr3_+_18002574 | 4.40 |
ENSMUST00000029080.5
|
Cypt12
|
cysteine-rich perinuclear theca 12 |
| chr3_-_15902583 | 4.22 |
ENSMUST00000108354.8
ENSMUST00000108349.2 ENSMUST00000108352.9 ENSMUST00000108350.8 ENSMUST00000050623.11 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr12_-_113790741 | 4.14 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr6_+_78347636 | 4.10 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr12_-_55061117 | 3.95 |
ENSMUST00000172875.8
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr5_+_29400981 | 3.89 |
ENSMUST00000160888.8
ENSMUST00000159272.8 ENSMUST00000001247.12 ENSMUST00000161398.8 ENSMUST00000160246.8 |
Rnf32
|
ring finger protein 32 |
| chr9_+_5298669 | 3.85 |
ENSMUST00000238505.2
|
Casp1
|
caspase 1 |
| chr7_-_24423715 | 3.78 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr6_-_69204417 | 3.77 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr10_+_87041814 | 3.68 |
ENSMUST00000189775.2
|
1700113H08Rik
|
RIKEN cDNA 1700113H08 gene |
| chr3_-_15491482 | 3.64 |
ENSMUST00000099201.9
ENSMUST00000194144.3 ENSMUST00000192700.3 |
Sirpb1a
|
signal-regulatory protein beta 1A |
| chr6_-_69394425 | 3.56 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chr6_-_40496403 | 3.54 |
ENSMUST00000031967.6
|
Prss37
|
protease, serine 37 |
| chr5_-_140986312 | 3.52 |
ENSMUST00000085786.7
|
Card11
|
caspase recruitment domain family, member 11 |
| chr6_-_68994064 | 3.49 |
ENSMUST00000103341.4
|
Igkv4-80
|
immunoglobulin kappa variable 4-80 |
| chr8_+_21382681 | 3.48 |
ENSMUST00000098908.4
|
Defb33
|
defensin beta 33 |
| chr6_+_78347844 | 3.42 |
ENSMUST00000096904.6
ENSMUST00000203266.2 |
Reg3b
|
regenerating islet-derived 3 beta |
| chr17_-_45970238 | 3.41 |
ENSMUST00000120717.8
|
Capn11
|
calpain 11 |
| chr6_-_69245427 | 3.33 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr6_+_68279392 | 3.31 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr19_+_8779903 | 3.24 |
ENSMUST00000172175.3
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr3_-_75072319 | 3.23 |
ENSMUST00000124618.2
|
Zbbx
|
zinc finger, B-box domain containing |
| chr5_+_107645626 | 3.19 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr3_+_104688363 | 3.17 |
ENSMUST00000002298.7
|
Ppm1j
|
protein phosphatase 1J |
| chr1_-_171854818 | 3.10 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr6_-_69162381 | 3.09 |
ENSMUST00000103344.3
|
Igkv4-74
|
immunoglobulin kappa variable 4-74 |
| chr1_-_28819331 | 3.08 |
ENSMUST00000059937.5
|
Gm597
|
predicted gene 597 |
| chr12_+_72488625 | 3.03 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr10_+_97482946 | 3.00 |
ENSMUST00000105285.4
|
Epyc
|
epiphycan |
| chr17_-_37399343 | 2.99 |
ENSMUST00000207101.2
ENSMUST00000217397.2 ENSMUST00000215195.2 ENSMUST00000216488.2 |
Olfr90
|
olfactory receptor 90 |
| chr10_-_53252210 | 2.87 |
ENSMUST00000095691.7
|
Cep85l
|
centrosomal protein 85-like |
| chr6_+_34006356 | 2.86 |
ENSMUST00000228187.2
ENSMUST00000070189.10 ENSMUST00000101564.9 |
Lrguk
|
leucine-rich repeats and guanylate kinase domain containing |
| chr9_+_40092216 | 2.82 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr1_-_162913210 | 2.81 |
ENSMUST00000096608.5
|
Mroh9
|
maestro heat-like repeat family member 9 |
| chr2_-_58050494 | 2.76 |
ENSMUST00000028175.7
|
Cytip
|
cytohesin 1 interacting protein |
| chr10_+_57527073 | 2.75 |
ENSMUST00000066028.13
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr6_-_130106861 | 2.64 |
ENSMUST00000014476.6
|
Klra8
|
killer cell lectin-like receptor, subfamily A, member 8 |
| chr8_+_84728123 | 2.63 |
ENSMUST00000060357.15
ENSMUST00000239176.2 |
1700067K01Rik
|
RIKEN cDNA 1700067K01 gene |
| chr8_-_79975199 | 2.59 |
ENSMUST00000034109.6
|
1700011L22Rik
|
RIKEN cDNA 1700011L22 gene |
| chr3_-_54962458 | 2.57 |
ENSMUST00000199352.2
ENSMUST00000198320.5 ENSMUST00000029368.7 |
Ccna1
|
cyclin A1 |
| chrX_+_111404963 | 2.56 |
ENSMUST00000026602.9
ENSMUST00000113412.3 |
2010106E10Rik
|
RIKEN cDNA 2010106E10 gene |
| chr2_+_118877594 | 2.51 |
ENSMUST00000152380.8
ENSMUST00000099542.9 |
Knl1
|
kinetochore scaffold 1 |
| chr4_+_147216495 | 2.50 |
ENSMUST00000084149.10
|
Zfp991
|
zinc finger protein 991 |
| chr1_+_139382485 | 2.49 |
ENSMUST00000200083.5
ENSMUST00000053364.12 |
Aspm
|
abnormal spindle microtubule assembly |
| chr19_-_11261177 | 2.47 |
ENSMUST00000186937.7
ENSMUST00000067673.13 |
Ms4a5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr4_+_146695418 | 2.41 |
ENSMUST00000130825.8
|
Zfp993
|
zinc finger protein 993 |
| chr7_+_130375799 | 2.39 |
ENSMUST00000048453.7
ENSMUST00000208593.2 |
Btbd16
|
BTB (POZ) domain containing 16 |
| chr10_+_57527084 | 2.39 |
ENSMUST00000175852.2
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr8_-_46542730 | 2.39 |
ENSMUST00000144244.2
|
Snx25
|
sorting nexin 25 |
| chrX_+_56257374 | 2.38 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chrX_+_110154017 | 2.36 |
ENSMUST00000210720.3
|
Cylc1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr4_+_52596266 | 2.36 |
ENSMUST00000029995.6
|
Toporsl
|
topoisomerase I binding, arginine/serine-rich like |
| chr15_-_9529898 | 2.34 |
ENSMUST00000228782.2
ENSMUST00000003981.6 |
Il7r
|
interleukin 7 receptor |
| chrX_-_56384089 | 2.34 |
ENSMUST00000033468.11
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr6_-_69020489 | 2.33 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chr2_+_118877610 | 2.33 |
ENSMUST00000153300.8
ENSMUST00000028799.12 |
Knl1
|
kinetochore scaffold 1 |
| chr6_-_68887922 | 2.32 |
ENSMUST00000103337.3
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr18_+_52748978 | 2.32 |
ENSMUST00000072666.4
ENSMUST00000209270.2 |
Zfp474
|
zinc finger protein 474 |
| chr4_+_145311759 | 2.32 |
ENSMUST00000119718.8
|
Zfp268
|
zinc finger protein 268 |
| chr3_-_16060545 | 2.25 |
ENSMUST00000194367.6
|
Gm5150
|
predicted gene 5150 |
| chr3_+_40755211 | 2.22 |
ENSMUST00000204473.2
|
Plk4
|
polo like kinase 4 |
| chr2_-_172212426 | 2.22 |
ENSMUST00000109139.8
ENSMUST00000028997.8 ENSMUST00000109140.10 |
Aurka
|
aurora kinase A |
| chr4_-_132072988 | 2.22 |
ENSMUST00000030726.13
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr8_-_86107593 | 2.19 |
ENSMUST00000122452.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr4_+_140427799 | 2.10 |
ENSMUST00000071169.9
|
Rcc2
|
regulator of chromosome condensation 2 |
| chr3_-_129834788 | 2.05 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr4_-_132073048 | 1.99 |
ENSMUST00000084250.11
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr5_-_62923463 | 1.95 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr9_-_20871081 | 1.95 |
ENSMUST00000177754.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr3_-_16060491 | 1.94 |
ENSMUST00000108347.3
|
Gm5150
|
predicted gene 5150 |
| chr6_-_69037208 | 1.91 |
ENSMUST00000103343.4
|
Igkv4-78
|
immunoglobulin kappa variable 4-78 |
| chrX_-_159689993 | 1.89 |
ENSMUST00000069417.6
|
Gja6
|
gap junction protein, alpha 6 |
| chr14_-_104760051 | 1.88 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr14_-_70666513 | 1.85 |
ENSMUST00000226426.2
ENSMUST00000048129.6 |
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chrX_+_106299484 | 1.85 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr3_-_86827664 | 1.82 |
ENSMUST00000194452.2
ENSMUST00000191752.6 |
Dclk2
|
doublecortin-like kinase 2 |
| chr10_-_111996226 | 1.82 |
ENSMUST00000220085.2
|
Glipr1l3
|
GLI pathogenesis-related 1 like 3 |
| chr7_+_101545547 | 1.82 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr1_-_33853598 | 1.80 |
ENSMUST00000019861.13
ENSMUST00000044455.8 |
Zfp451
|
zinc finger protein 451 |
| chr19_-_32173824 | 1.79 |
ENSMUST00000151822.2
|
Sgms1
|
sphingomyelin synthase 1 |
| chr6_-_130170075 | 1.73 |
ENSMUST00000112032.8
ENSMUST00000071554.3 |
Klra9
|
killer cell lectin-like receptor subfamily A, member 9 |
| chr4_+_145311722 | 1.73 |
ENSMUST00000105739.8
|
Zfp268
|
zinc finger protein 268 |
| chr2_-_45007407 | 1.72 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr6_+_129554868 | 1.64 |
ENSMUST00000053708.9
|
Klre1
|
killer cell lectin-like receptor family E member 1 |
| chr19_-_53932581 | 1.55 |
ENSMUST00000236885.2
ENSMUST00000236098.2 ENSMUST00000236370.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr11_-_99482165 | 1.52 |
ENSMUST00000104930.2
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr5_-_65855511 | 1.50 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr19_-_53932867 | 1.48 |
ENSMUST00000235688.2
ENSMUST00000235348.2 |
Bbip1
|
BBSome interacting protein 1 |
| chrX_+_16485937 | 1.47 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr6_-_129484070 | 1.43 |
ENSMUST00000183258.8
ENSMUST00000182784.4 ENSMUST00000032265.13 ENSMUST00000162815.2 |
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr2_+_87696836 | 1.41 |
ENSMUST00000213308.3
|
Olfr1152
|
olfactory receptor 1152 |
| chr18_+_67597929 | 1.40 |
ENSMUST00000025411.9
|
Prelid3a
|
PRELI domain containing 3A |
| chr3_-_86827640 | 1.39 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr6_-_68968278 | 1.39 |
ENSMUST00000197966.2
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chr7_+_89814713 | 1.38 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr14_+_122272069 | 1.38 |
ENSMUST00000045976.7
|
Timm8a2
|
translocase of inner mitochondrial membrane 8A2 |
| chr13_-_103042294 | 1.35 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chrX_-_111315519 | 1.35 |
ENSMUST00000124335.8
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr4_-_117539431 | 1.35 |
ENSMUST00000102687.4
|
Dmap1
|
DNA methyltransferase 1-associated protein 1 |
| chr13_-_43634695 | 1.34 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr6_+_123239076 | 1.34 |
ENSMUST00000032240.4
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr6_+_8948608 | 1.33 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr2_+_69652714 | 1.31 |
ENSMUST00000053087.4
|
Klhl23
|
kelch-like 23 |
| chr3_-_54962922 | 1.27 |
ENSMUST00000197238.5
|
Ccna1
|
cyclin A1 |
| chr2_+_127696548 | 1.24 |
ENSMUST00000028859.8
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr10_-_112764879 | 1.21 |
ENSMUST00000099276.4
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr16_-_29363671 | 1.21 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr15_-_79658584 | 1.21 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr2_-_111843053 | 1.21 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr13_-_103042554 | 1.20 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr4_+_102427247 | 1.18 |
ENSMUST00000097950.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr12_+_112727089 | 1.17 |
ENSMUST00000063888.5
|
Pld4
|
phospholipase D family, member 4 |
| chr11_-_99121822 | 1.17 |
ENSMUST00000103133.4
|
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr1_-_144427302 | 1.16 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr18_-_24736521 | 1.15 |
ENSMUST00000154205.2
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr14_-_54651442 | 1.15 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr3_-_54962899 | 1.14 |
ENSMUST00000199144.5
|
Ccna1
|
cyclin A1 |
| chr1_+_65225834 | 1.13 |
ENSMUST00000081154.14
|
Pikfyve
|
phosphoinositide kinase, FYVE type zinc finger containing |
| chr15_-_79658608 | 1.12 |
ENSMUST00000229644.2
ENSMUST00000023055.8 |
Dnal4
|
dynein, axonemal, light chain 4 |
| chr2_-_164427367 | 1.12 |
ENSMUST00000109342.2
|
Wfdc6a
|
WAP four-disulfide core domain 6A |
| chr4_-_43710231 | 1.10 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr5_-_140687995 | 1.09 |
ENSMUST00000135028.5
ENSMUST00000077890.12 ENSMUST00000041783.14 ENSMUST00000142081.6 |
Iqce
|
IQ motif containing E |
| chr14_+_62529924 | 1.08 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr19_-_24178000 | 1.07 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr13_+_51799268 | 1.07 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr4_-_147894245 | 1.07 |
ENSMUST00000105734.10
ENSMUST00000176201.2 |
Zfp984
Gm20707
|
zinc finger protein 984 predicted gene 20707 |
| chr2_-_111880531 | 1.04 |
ENSMUST00000213582.2
ENSMUST00000213961.3 ENSMUST00000215531.2 |
Olfr1312
|
olfactory receptor 1312 |
| chr11_-_99494134 | 1.02 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr4_+_146599377 | 1.02 |
ENSMUST00000140089.7
ENSMUST00000179175.3 |
Zfp981
|
zinc finger protein 981 |
| chr1_+_190660689 | 1.00 |
ENSMUST00000066632.14
ENSMUST00000110899.7 |
Angel2
|
angel homolog 2 |
| chrX_+_110808231 | 1.00 |
ENSMUST00000207546.2
|
Gm45194
|
predicted gene 45194 |
| chr18_+_37898633 | 0.98 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr4_+_134658209 | 0.97 |
ENSMUST00000030622.3
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chr14_+_32043944 | 0.95 |
ENSMUST00000022480.8
ENSMUST00000228529.2 |
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr6_-_69261303 | 0.93 |
ENSMUST00000103349.2
|
Igkv4-69
|
immunoglobulin kappa variable 4-69 |
| chr10_+_80003612 | 0.93 |
ENSMUST00000105365.9
|
Cirbp
|
cold inducible RNA binding protein |
| chr6_-_130314465 | 0.89 |
ENSMUST00000088017.5
ENSMUST00000111998.9 |
Klra3
|
killer cell lectin-like receptor, subfamily A, member 3 |
| chr4_+_132495636 | 0.88 |
ENSMUST00000102561.11
|
Rpa2
|
replication protein A2 |
| chr6_-_69377328 | 0.87 |
ENSMUST00000198345.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr15_-_8740218 | 0.83 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr3_+_32490300 | 0.83 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr1_+_40478926 | 0.83 |
ENSMUST00000173514.8
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr2_+_121787131 | 0.81 |
ENSMUST00000110574.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr17_-_56343531 | 0.81 |
ENSMUST00000233803.2
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr1_+_65225886 | 0.80 |
ENSMUST00000097707.5
|
Pikfyve
|
phosphoinositide kinase, FYVE type zinc finger containing |
| chr13_+_21901791 | 0.80 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10 |
| chr7_-_11414074 | 0.80 |
ENSMUST00000227010.2
ENSMUST00000209638.3 |
Vmn1r72
|
vomeronasal 1 receptor 72 |
| chr5_+_140593075 | 0.80 |
ENSMUST00000031555.3
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_-_4479445 | 0.79 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr17_-_40553176 | 0.79 |
ENSMUST00000026499.6
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr4_+_146586445 | 0.78 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr3_-_96359622 | 0.78 |
ENSMUST00000093126.11
ENSMUST00000098841.4 |
BC107364
|
cDNA sequence BC107364 |
| chr14_-_36820304 | 0.78 |
ENSMUST00000022337.11
|
Cdhr1
|
cadherin-related family member 1 |
| chr1_+_40478787 | 0.77 |
ENSMUST00000097772.10
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr4_+_109092610 | 0.76 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr19_-_53933052 | 0.76 |
ENSMUST00000135402.4
|
Bbip1
|
BBSome interacting protein 1 |
| chr2_+_88470886 | 0.75 |
ENSMUST00000217379.2
ENSMUST00000120598.3 |
Olfr1191-ps1
|
olfactory receptor 1191, pseudogene 1 |
| chr16_-_55755208 | 0.74 |
ENSMUST00000121129.8
ENSMUST00000023270.14 |
Cep97
|
centrosomal protein 97 |
| chr16_-_55755160 | 0.74 |
ENSMUST00000122280.8
ENSMUST00000121703.3 |
Cep97
|
centrosomal protein 97 |
| chr4_+_145397238 | 0.72 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chrX_-_88976458 | 0.71 |
ENSMUST00000088146.3
|
4930415L06Rik
|
RIKEN cDNA 4930415L06 gene |
| chr2_-_17465410 | 0.71 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr8_+_46463633 | 0.70 |
ENSMUST00000110381.9
|
Lrp2bp
|
Lrp2 binding protein |
| chr17_-_56343625 | 0.70 |
ENSMUST00000003268.11
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr13_+_43019718 | 0.70 |
ENSMUST00000131942.2
|
Phactr1
|
phosphatase and actin regulator 1 |
| chrX_-_8391193 | 0.69 |
ENSMUST00000115573.2
|
Gm14459
|
predicted gene 14459 |
| chr11_-_76386190 | 0.69 |
ENSMUST00000108408.9
|
Abr
|
active BCR-related gene |
| chr17_+_37148015 | 0.68 |
ENSMUST00000179968.8
ENSMUST00000130367.8 ENSMUST00000053434.15 ENSMUST00000130801.8 ENSMUST00000144182.8 ENSMUST00000123715.8 |
Trim26
|
tripartite motif-containing 26 |
| chr2_-_88157559 | 0.68 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr7_+_101546059 | 0.68 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr1_+_161796748 | 0.66 |
ENSMUST00000111594.3
ENSMUST00000028021.7 ENSMUST00000193784.2 ENSMUST00000161826.2 |
Pigc
Gm38304
|
phosphatidylinositol glycan anchor biosynthesis, class C predicted gene, 38304 |
| chr2_-_73410632 | 0.66 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr13_-_74956030 | 0.66 |
ENSMUST00000065629.6
|
Cast
|
calpastatin |
| chr11_+_43572825 | 0.65 |
ENSMUST00000061070.6
ENSMUST00000094294.5 |
Pwwp2a
|
PWWP domain containing 2A |
| chr10_+_115854118 | 0.65 |
ENSMUST00000063470.11
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr9_+_105272507 | 0.63 |
ENSMUST00000035181.10
ENSMUST00000123807.8 |
Aste1
|
asteroid homolog 1 |
| chr2_-_79738773 | 0.62 |
ENSMUST00000102652.10
ENSMUST00000102651.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr18_+_59195534 | 0.59 |
ENSMUST00000058633.9
ENSMUST00000175897.8 ENSMUST00000118510.8 ENSMUST00000175830.2 |
Minar2
|
membrane integral NOTCH2 associated receptor 2 |
| chr9_+_39932760 | 0.58 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr10_+_115979787 | 0.58 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr4_+_109092459 | 0.58 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 16.0 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.0 | 3.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 1.0 | 3.8 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.8 | 2.4 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.7 | 2.2 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.7 | 2.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.6 | 7.5 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.5 | 2.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.5 | 7.0 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.5 | 1.9 | GO:0090309 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.5 | 1.9 | GO:0000239 | pachytene(GO:0000239) |
| 0.4 | 8.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.4 | 2.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.4 | 7.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.4 | 1.6 | GO:0002859 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.4 | 1.6 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.4 | 2.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.4 | 2.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.3 | 4.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.3 | 2.2 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.3 | 0.8 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.3 | 1.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.3 | 1.8 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.2 | 1.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 5.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 64.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 1.4 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 1.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 3.8 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 | 1.3 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 2.3 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 1.5 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 1.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 0.3 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 | 0.8 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.2 | 1.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 1.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 2.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.6 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.4 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 3.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 2.5 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 1.5 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.1 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.1 | 0.5 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.1 | 0.5 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.1 | 15.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.2 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.9 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 4.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 1.5 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 2.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 1.4 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.3 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 3.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.6 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 1.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 1.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 3.6 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.1 | 1.9 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.3 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 1.9 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 1.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.4 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.7 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 1.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 1.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 2.9 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0015691 | vanadium ion transport(GO:0015676) cadmium ion transport(GO:0015691) lead ion transport(GO:0015692) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 1.8 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.3 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 4.0 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.4 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 1.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 5.0 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 1.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 1.3 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.0 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 1.0 | 3.1 | GO:0060187 | cell pole(GO:0060187) |
| 1.0 | 4.0 | GO:0008623 | CHRAC(GO:0008623) |
| 1.0 | 3.8 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.6 | 2.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.4 | 4.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.4 | 1.9 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.3 | 2.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 3.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 0.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.3 | 2.9 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 8.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 1.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 1.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 6.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 15.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 4.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 2.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 3.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 2.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 1.0 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 2.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 3.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.9 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.9 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 2.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 69.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 10.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 3.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 5.7 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 3.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 4.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.8 | 2.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.8 | 8.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.6 | 7.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.6 | 4.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.6 | 7.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.5 | 1.6 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 1.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.3 | 1.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 1.8 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 0.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 1.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 0.7 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 2.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 1.9 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 5.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 1.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 1.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 1.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 1.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 1.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 2.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 17.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.5 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 3.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 1.0 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.6 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 8.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.9 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 2.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.8 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 0.8 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.9 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 13.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 1.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 1.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 1.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 7.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.7 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 2.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 2.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 2.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 2.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 2.7 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 2.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 2.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 8.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 4.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 3.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 2.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 2.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 3.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 3.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 3.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 4.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 2.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 6.5 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 2.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 2.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 5.1 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.2 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 2.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 2.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |