Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Hoxd11_Cdx1_Hoxc11
Z-value: 1.70
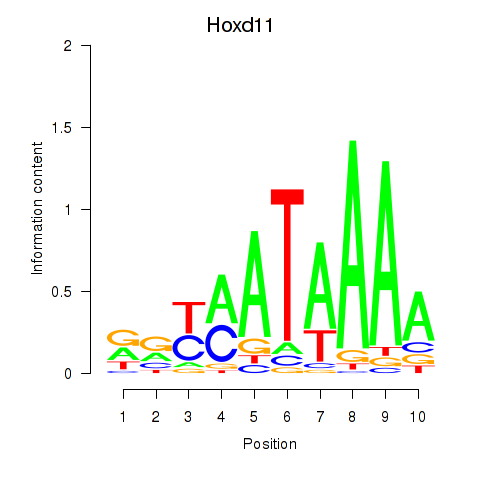
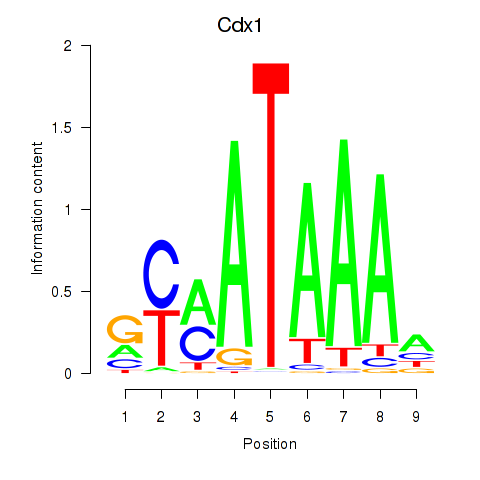
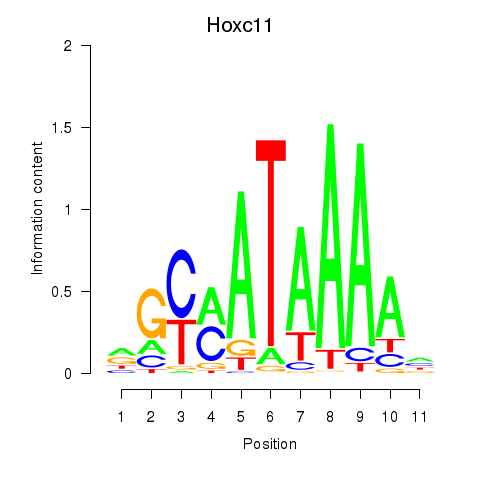
Transcription factors associated with Hoxd11_Cdx1_Hoxc11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd11
|
ENSMUSG00000042499.14 | Hoxd11 |
|
Cdx1
|
ENSMUSG00000024619.9 | Cdx1 |
|
Hoxc11
|
ENSMUSG00000001656.4 | Hoxc11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cdx1 | mm39_v1_chr18_-_61169262_61169282 | 0.74 | 8.1e-14 | Click! |
| Hoxd11 | mm39_v1_chr2_+_74512638_74512670 | 0.43 | 1.6e-04 | Click! |
| Hoxc11 | mm39_v1_chr15_+_102862862_102862931 | 0.10 | 3.9e-01 | Click! |
Activity profile of Hoxd11_Cdx1_Hoxc11 motif
Sorted Z-values of Hoxd11_Cdx1_Hoxc11 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd11_Cdx1_Hoxc11
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_71176811 | 30.18 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr7_-_135130374 | 24.68 |
ENSMUST00000053716.8
|
Clrn3
|
clarin 3 |
| chr3_+_3699205 | 23.35 |
ENSMUST00000108394.3
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr4_+_11758147 | 22.90 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chrX_+_95498965 | 20.47 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr11_-_100036792 | 17.57 |
ENSMUST00000007317.8
|
Krt19
|
keratin 19 |
| chr7_+_123061497 | 16.71 |
ENSMUST00000033023.10
|
Aqp8
|
aquaporin 8 |
| chr13_-_24098981 | 16.66 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr7_+_123061535 | 16.61 |
ENSMUST00000098056.6
|
Aqp8
|
aquaporin 8 |
| chr17_+_31652073 | 16.46 |
ENSMUST00000237363.2
|
Pde9a
|
phosphodiesterase 9A |
| chr17_+_31652029 | 16.42 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr16_-_56616186 | 15.86 |
ENSMUST00000023437.5
|
Adgrg7
|
adhesion G protein-coupled receptor G7 |
| chr7_-_140856642 | 15.82 |
ENSMUST00000080654.7
ENSMUST00000167263.9 |
Cdhr5
|
cadherin-related family member 5 |
| chr13_-_24098951 | 15.79 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr14_-_36832044 | 14.20 |
ENSMUST00000179488.3
|
2610528A11Rik
|
RIKEN cDNA 2610528A11 gene |
| chr10_-_116732813 | 13.35 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr10_+_116013256 | 13.34 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr3_-_144638284 | 12.25 |
ENSMUST00000098549.4
|
Clca4b
|
chloride channel accessory 4B |
| chr2_+_57887896 | 11.55 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr2_+_163348728 | 11.08 |
ENSMUST00000143911.8
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr1_+_165958036 | 10.69 |
ENSMUST00000166860.2
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr10_+_116013122 | 10.67 |
ENSMUST00000148731.8
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr8_-_5155347 | 9.97 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr3_-_72875187 | 9.20 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr17_-_43813664 | 9.03 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr6_-_24168082 | 8.81 |
ENSMUST00000031713.9
|
Slc13a1
|
solute carrier family 13 (sodium/sulfate symporters), member 1 |
| chr19_-_10995390 | 8.59 |
ENSMUST00000177684.3
|
Ms4a18
|
membrane-spanning 4-domains, subfamily A, member 18 |
| chr11_+_96085118 | 7.77 |
ENSMUST00000062709.4
|
Hoxb13
|
homeobox B13 |
| chr7_+_130633776 | 7.59 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chrX_+_138464065 | 7.35 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr6_-_52203146 | 7.25 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr15_-_78739717 | 7.15 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr3_+_57332735 | 7.00 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr3_-_144555062 | 6.92 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr19_+_24853039 | 6.89 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr18_+_21205386 | 6.88 |
ENSMUST00000082235.5
|
Mep1b
|
meprin 1 beta |
| chr2_-_103114105 | 6.58 |
ENSMUST00000111174.8
|
Ehf
|
ets homologous factor |
| chr18_-_32170012 | 6.43 |
ENSMUST00000134663.2
|
Myo7b
|
myosin VIIB |
| chr1_+_165957784 | 6.41 |
ENSMUST00000060833.14
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr7_+_141276575 | 6.40 |
ENSMUST00000185406.8
|
Muc2
|
mucin 2 |
| chr11_-_43492367 | 6.09 |
ENSMUST00000020672.5
|
Fabp6
|
fatty acid binding protein 6 |
| chr3_-_63758672 | 6.05 |
ENSMUST00000162269.9
ENSMUST00000159676.9 ENSMUST00000175947.8 |
Plch1
|
phospholipase C, eta 1 |
| chr19_+_56414114 | 5.56 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chr10_+_79656823 | 5.49 |
ENSMUST00000169041.9
|
Misp
|
mitotic spindle positioning |
| chrX_+_162923474 | 5.47 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr3_+_138019040 | 5.40 |
ENSMUST00000013455.13
ENSMUST00000106247.2 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr16_-_21814190 | 5.08 |
ENSMUST00000231766.2
ENSMUST00000074230.12 |
Liph
|
lipase, member H |
| chr1_+_165957909 | 4.75 |
ENSMUST00000166159.2
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr2_+_122129379 | 4.62 |
ENSMUST00000028656.2
|
Duoxa2
|
dual oxidase maturation factor 2 |
| chr6_-_83513222 | 4.59 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr6_-_52195663 | 4.54 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7 |
| chr1_-_45542442 | 4.49 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr6_-_83513184 | 4.33 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr1_+_74752710 | 4.32 |
ENSMUST00000027356.7
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr3_+_62327089 | 4.31 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr7_-_30312246 | 4.28 |
ENSMUST00000006476.6
|
Upk1a
|
uroplakin 1A |
| chr16_-_21814289 | 4.24 |
ENSMUST00000060673.8
|
Liph
|
lipase, member H |
| chr17_+_3447465 | 4.20 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr9_-_9239056 | 4.18 |
ENSMUST00000093893.12
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr3_+_138121245 | 3.91 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr1_-_128256048 | 3.88 |
ENSMUST00000073490.7
|
Lct
|
lactase |
| chr13_-_23806530 | 3.83 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr10_+_5589210 | 3.83 |
ENSMUST00000019906.6
|
Vip
|
vasoactive intestinal polypeptide |
| chr5_+_63969706 | 3.78 |
ENSMUST00000081747.8
ENSMUST00000196575.5 |
0610040J01Rik
|
RIKEN cDNA 0610040J01 gene |
| chr2_-_160208977 | 3.68 |
ENSMUST00000099126.5
|
Mafb
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
| chr6_-_52211882 | 3.62 |
ENSMUST00000125581.2
|
Hoxa10
|
homeobox A10 |
| chr2_+_15531281 | 3.41 |
ENSMUST00000146205.3
|
Malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr1_-_57008986 | 3.38 |
ENSMUST00000176759.2
ENSMUST00000177424.2 |
Satb2
|
special AT-rich sequence binding protein 2 |
| chr7_-_19411866 | 3.20 |
ENSMUST00000142352.9
|
Apoc2
|
apolipoprotein C-II |
| chr1_-_37535170 | 3.11 |
ENSMUST00000148047.2
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr1_-_37535202 | 3.07 |
ENSMUST00000143636.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr5_-_87485023 | 3.05 |
ENSMUST00000031195.3
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr1_+_177557380 | 3.05 |
ENSMUST00000016106.6
|
1700016C15Rik
|
RIKEN cDNA 1700016C15 gene |
| chr6_-_70412460 | 3.02 |
ENSMUST00000103394.2
|
Igkv6-14
|
immunoglobulin kappa variable 6-14 |
| chr10_-_40018243 | 2.99 |
ENSMUST00000092566.8
ENSMUST00000213488.2 |
Slc16a10
|
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
| chr6_-_119365632 | 2.98 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr5_-_120610828 | 2.97 |
ENSMUST00000052258.14
ENSMUST00000031594.13 |
Sdsl
|
serine dehydratase-like |
| chr14_+_31807760 | 2.97 |
ENSMUST00000170600.8
ENSMUST00000168986.7 ENSMUST00000169649.2 |
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr10_+_101517348 | 2.94 |
ENSMUST00000179929.8
ENSMUST00000219195.2 ENSMUST00000127504.9 |
Mgat4c
|
MGAT4 family, member C |
| chr3_-_88366351 | 2.57 |
ENSMUST00000165898.8
ENSMUST00000127436.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr5_-_73349191 | 2.56 |
ENSMUST00000176910.3
|
Fryl
|
FRY like transcription coactivator |
| chrX_-_133442596 | 2.54 |
ENSMUST00000054213.5
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chr3_-_144525255 | 2.53 |
ENSMUST00000029929.12
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr9_+_103917821 | 2.49 |
ENSMUST00000216593.2
ENSMUST00000147249.3 |
Nphp3
Gm28305
|
nephronophthisis 3 (adolescent) predicted gene 28305 |
| chr8_+_110717062 | 2.46 |
ENSMUST00000001720.14
ENSMUST00000143741.2 |
Tat
|
tyrosine aminotransferase |
| chr15_-_101336669 | 2.44 |
ENSMUST00000081945.5
|
Krt87
|
keratin 87 |
| chr6_-_70435020 | 2.44 |
ENSMUST00000198184.2
|
Igkv6-13
|
immunoglobulin kappa variable 6-13 |
| chr1_-_71692320 | 2.41 |
ENSMUST00000186940.7
ENSMUST00000188894.7 ENSMUST00000188674.7 ENSMUST00000189821.7 ENSMUST00000187938.7 ENSMUST00000190780.7 ENSMUST00000186736.2 ENSMUST00000055226.13 ENSMUST00000186129.7 |
Fn1
|
fibronectin 1 |
| chr4_-_102883905 | 2.40 |
ENSMUST00000084382.6
ENSMUST00000106869.3 |
Insl5
|
insulin-like 5 |
| chr3_-_49711706 | 2.37 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chrX_+_55824797 | 2.37 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr19_+_58748132 | 2.29 |
ENSMUST00000026081.5
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr9_+_110948492 | 2.23 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr17_-_35293447 | 2.21 |
ENSMUST00000007259.4
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr5_-_51725059 | 2.16 |
ENSMUST00000127135.3
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr17_+_71326510 | 2.16 |
ENSMUST00000073211.13
ENSMUST00000024847.14 |
Myom1
|
myomesin 1 |
| chr8_+_57774010 | 2.11 |
ENSMUST00000040104.5
|
Hand2
|
heart and neural crest derivatives expressed 2 |
| chr10_-_128996851 | 2.04 |
ENSMUST00000078914.3
|
Olfr771
|
olfactory receptor 771 |
| chr7_+_126808016 | 2.04 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr17_-_36207965 | 1.98 |
ENSMUST00000150056.2
ENSMUST00000156817.2 ENSMUST00000146451.8 ENSMUST00000148482.8 |
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr17_+_71326542 | 1.97 |
ENSMUST00000179759.3
|
Myom1
|
myomesin 1 |
| chr1_-_37575313 | 1.97 |
ENSMUST00000042161.15
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr9_+_111014254 | 1.92 |
ENSMUST00000198986.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr1_+_45350698 | 1.90 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr10_-_107321938 | 1.89 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr3_-_49711765 | 1.86 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr10_+_26698556 | 1.82 |
ENSMUST00000135866.2
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr1_-_144052997 | 1.81 |
ENSMUST00000111941.2
ENSMUST00000052375.8 |
Rgs13
|
regulator of G-protein signaling 13 |
| chr1_-_144124871 | 1.79 |
ENSMUST00000189061.7
|
Rgs1
|
regulator of G-protein signaling 1 |
| chrX_+_132809189 | 1.76 |
ENSMUST00000113304.2
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr10_-_25076008 | 1.71 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr2_-_136727599 | 1.70 |
ENSMUST00000231720.2
|
ENSMUSG00000116563.2
|
novel protein |
| chr17_-_16046780 | 1.64 |
ENSMUST00000232638.2
ENSMUST00000170578.3 |
Rgmb
|
repulsive guidance molecule family member B |
| chr6_-_124942457 | 1.63 |
ENSMUST00000112439.9
|
Cops7a
|
COP9 signalosome subunit 7A |
| chr6_-_124942366 | 1.63 |
ENSMUST00000129446.8
ENSMUST00000032220.15 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr4_+_116578117 | 1.62 |
ENSMUST00000045542.13
ENSMUST00000106459.8 |
Tesk2
|
testis-specific kinase 2 |
| chr15_+_25774070 | 1.62 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr3_-_19319123 | 1.59 |
ENSMUST00000121951.2
|
Pde7a
|
phosphodiesterase 7A |
| chr7_-_44711771 | 1.57 |
ENSMUST00000210101.2
ENSMUST00000209219.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr8_-_45747883 | 1.54 |
ENSMUST00000026907.6
|
Klkb1
|
kallikrein B, plasma 1 |
| chr4_-_76262464 | 1.50 |
ENSMUST00000050757.16
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr5_+_4073343 | 1.48 |
ENSMUST00000238634.2
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr3_-_88366159 | 1.46 |
ENSMUST00000147200.8
ENSMUST00000169222.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr8_-_70355208 | 1.45 |
ENSMUST00000110167.5
|
Ndufa13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr18_+_65054188 | 1.45 |
ENSMUST00000236898.2
ENSMUST00000237410.2 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr11_-_21320452 | 1.45 |
ENSMUST00000102875.11
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr15_+_59186876 | 1.45 |
ENSMUST00000022977.14
ENSMUST00000100640.5 |
Sqle
|
squalene epoxidase |
| chr7_-_141015240 | 1.45 |
ENSMUST00000138865.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr2_+_113235475 | 1.44 |
ENSMUST00000238883.2
|
Fmn1
|
formin 1 |
| chr5_-_73413888 | 1.43 |
ENSMUST00000101127.12
|
Fryl
|
FRY like transcription coactivator |
| chr8_-_58106057 | 1.41 |
ENSMUST00000034021.12
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr2_-_86180622 | 1.39 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr9_-_96774719 | 1.36 |
ENSMUST00000154146.8
|
Pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr7_-_44711075 | 1.34 |
ENSMUST00000007981.9
ENSMUST00000210500.2 ENSMUST00000210493.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr7_+_140343652 | 1.32 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr10_-_105410280 | 1.28 |
ENSMUST00000061506.9
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr14_-_31807552 | 1.23 |
ENSMUST00000022461.11
ENSMUST00000067955.12 ENSMUST00000124303.9 |
Dph3
|
diphthamine biosynthesis 3 |
| chr9_-_44710480 | 1.20 |
ENSMUST00000214833.2
ENSMUST00000213972.2 ENSMUST00000214431.2 ENSMUST00000213363.2 ENSMUST00000114705.9 ENSMUST00000002100.8 |
Tmem25
|
transmembrane protein 25 |
| chr13_+_13612136 | 1.17 |
ENSMUST00000005532.9
|
Nid1
|
nidogen 1 |
| chr7_-_44711130 | 1.16 |
ENSMUST00000211337.2
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr16_-_55659194 | 1.16 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr2_-_32341408 | 1.14 |
ENSMUST00000028160.15
ENSMUST00000113310.9 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr9_+_21746785 | 1.13 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr14_+_28233301 | 1.13 |
ENSMUST00000112272.2
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr6_-_124942170 | 1.09 |
ENSMUST00000148485.2
ENSMUST00000129976.8 |
Cops7a
|
COP9 signalosome subunit 7A |
| chrX_-_7185424 | 1.09 |
ENSMUST00000115746.8
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr19_-_7943365 | 1.08 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr3_+_102981352 | 1.06 |
ENSMUST00000176440.2
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr11_+_97689819 | 1.04 |
ENSMUST00000143571.2
|
Lasp1
|
LIM and SH3 protein 1 |
| chr4_-_49408040 | 1.01 |
ENSMUST00000081541.9
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr2_-_140513382 | 0.98 |
ENSMUST00000110057.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr4_-_63965161 | 0.97 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr12_-_111933328 | 0.97 |
ENSMUST00000222874.2
ENSMUST00000223191.2 ENSMUST00000021719.7 |
Atp5mpl
|
ATP synthase membrane subunit 6.8PL |
| chr5_-_87572060 | 0.94 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chrX_+_132809166 | 0.93 |
ENSMUST00000033606.15
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr7_+_127399776 | 0.91 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr11_-_101785181 | 0.88 |
ENSMUST00000057054.8
|
Meox1
|
mesenchyme homeobox 1 |
| chr14_-_24053994 | 0.87 |
ENSMUST00000225431.2
ENSMUST00000188210.8 ENSMUST00000224787.2 ENSMUST00000225315.2 ENSMUST00000225556.2 ENSMUST00000223727.2 ENSMUST00000223655.2 ENSMUST00000224077.2 ENSMUST00000224812.2 ENSMUST00000224285.2 ENSMUST00000225471.2 ENSMUST00000224232.2 ENSMUST00000223749.2 ENSMUST00000224025.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr2_-_140513320 | 0.87 |
ENSMUST00000056760.4
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr15_-_77129786 | 0.84 |
ENSMUST00000228558.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr15_-_77129706 | 0.83 |
ENSMUST00000228361.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr3_+_82915031 | 0.83 |
ENSMUST00000048486.13
ENSMUST00000194175.2 |
Fgg
|
fibrinogen gamma chain |
| chr15_-_34356567 | 0.82 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr11_-_17161504 | 0.82 |
ENSMUST00000020317.8
|
Pno1
|
partner of NOB1 homolog |
| chr16_+_88812172 | 0.82 |
ENSMUST00000089106.4
|
Gm10229
|
predicted gene 10229 |
| chr16_+_29884153 | 0.82 |
ENSMUST00000023171.8
|
Hes1
|
hes family bHLH transcription factor 1 |
| chr19_-_6117815 | 0.81 |
ENSMUST00000162575.8
ENSMUST00000159084.8 ENSMUST00000161718.8 ENSMUST00000162810.8 ENSMUST00000025713.12 ENSMUST00000113543.9 ENSMUST00000160417.8 ENSMUST00000161528.2 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr13_+_81031512 | 0.81 |
ENSMUST00000099356.10
|
Arrdc3
|
arrestin domain containing 3 |
| chr16_+_41353360 | 0.81 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr19_-_7319211 | 0.81 |
ENSMUST00000171393.8
|
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr6_-_139478919 | 0.79 |
ENSMUST00000170650.3
|
Rergl
|
RERG/RAS-like |
| chr2_-_26012751 | 0.77 |
ENSMUST00000140993.2
ENSMUST00000028300.6 |
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chrX_-_7185529 | 0.76 |
ENSMUST00000128319.2
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr7_-_141014477 | 0.75 |
ENSMUST00000106007.10
ENSMUST00000150026.2 ENSMUST00000202840.4 ENSMUST00000133206.9 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr4_+_118217179 | 0.74 |
ENSMUST00000006562.6
|
Hyi
|
hydroxypyruvate isomerase (putative) |
| chr19_+_44481901 | 0.72 |
ENSMUST00000041163.5
|
Wnt8b
|
wingless-type MMTV integration site family, member 8B |
| chr17_-_36208265 | 0.71 |
ENSMUST00000148721.8
|
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr2_+_29678248 | 0.69 |
ENSMUST00000028137.10
ENSMUST00000148791.2 |
Coq4
|
coenzyme Q4 |
| chr17_+_8502682 | 0.69 |
ENSMUST00000124023.8
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr12_+_55349422 | 0.69 |
ENSMUST00000021411.15
|
Prorp
|
protein only RNase P catalytic subunit |
| chr10_-_30494333 | 0.68 |
ENSMUST00000019925.7
|
Hint3
|
histidine triad nucleotide binding protein 3 |
| chr15_-_41733099 | 0.67 |
ENSMUST00000054742.7
|
Abra
|
actin-binding Rho activating protein |
| chr18_-_47466378 | 0.67 |
ENSMUST00000126684.2
ENSMUST00000156422.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr13_+_46427176 | 0.67 |
ENSMUST00000076622.4
|
Stmnd1
|
stathmin domain containing 1 |
| chr10_-_53952686 | 0.67 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr10_+_23814470 | 0.67 |
ENSMUST00000079134.5
|
Taar2
|
trace amine-associated receptor 2 |
| chr13_+_22220000 | 0.65 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12 |
| chr16_-_58459102 | 0.64 |
ENSMUST00000053249.9
|
Ftdc2
|
ferritin domain containing 2 |
| chr14_-_7750938 | 0.63 |
ENSMUST00000223740.3
ENSMUST00000224049.3 ENSMUST00000224672.3 ENSMUST00000224752.3 ENSMUST00000223695.3 ENSMUST00000224333.3 ENSMUST00000224952.3 |
Slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr15_-_101422054 | 0.63 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chr2_+_74552322 | 0.63 |
ENSMUST00000047904.4
|
Hoxd4
|
homeobox D4 |
| chrX_+_81992467 | 0.63 |
ENSMUST00000114000.8
|
Dmd
|
dystrophin, muscular dystrophy |
| chr19_-_45224251 | 0.63 |
ENSMUST00000099401.6
|
Lbx1
|
ladybird homeobox 1 |
| chr13_-_3995349 | 0.61 |
ENSMUST00000058610.8
|
Ucn3
|
urocortin 3 |
| chr3_+_122298308 | 0.60 |
ENSMUST00000199358.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr9_-_101076198 | 0.58 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr16_-_35589726 | 0.58 |
ENSMUST00000023554.9
|
Slc49a4
|
solute carrier family 49 member 4 |
| chr3_+_89366425 | 0.56 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase |
| chr2_+_145627900 | 0.55 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr1_-_144124787 | 0.55 |
ENSMUST00000185714.2
|
Rgs1
|
regulator of G-protein signaling 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 22.9 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 4.4 | 22.2 | GO:1904970 | brush border assembly(GO:1904970) |
| 4.2 | 33.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 4.0 | 24.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 3.7 | 11.1 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 1.9 | 7.8 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 1.9 | 30.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 1.5 | 32.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 1.4 | 5.5 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 1.1 | 4.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.1 | 5.6 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 1.0 | 3.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.8 | 4.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.8 | 4.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.8 | 2.4 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.8 | 3.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.7 | 3.7 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.7 | 3.7 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.7 | 2.2 | GO:1903210 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.6 | 1.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.6 | 4.6 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.6 | 3.4 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.5 | 19.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.5 | 1.5 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.4 | 2.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.4 | 0.8 | GO:2000974 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.4 | 8.8 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.4 | 1.2 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.4 | 10.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.4 | 1.5 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.4 | 6.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.4 | 1.1 | GO:0061354 | chemorepulsion of dopaminergic neuron axon(GO:0036518) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) melanocyte proliferation(GO:0097325) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.4 | 2.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) maintenance of organ identity(GO:0048496) |
| 0.4 | 17.6 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.3 | 4.5 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.3 | 22.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.3 | 10.0 | GO:0060065 | uterus development(GO:0060065) |
| 0.3 | 3.8 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.3 | 7.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 1.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 1.7 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.3 | 2.5 | GO:0046689 | tyrosine catabolic process(GO:0006572) response to mercury ion(GO:0046689) |
| 0.3 | 0.5 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.3 | 0.8 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.3 | 1.8 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.3 | 4.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 0.8 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.2 | 3.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 1.9 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 4.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 0.6 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.2 | 33.3 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.2 | 1.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 2.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 1.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 | 3.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 3.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.4 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.2 | 6.1 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.2 | 3.0 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 2.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 7.4 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.2 | 4.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.9 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.1 | 0.4 | GO:0014732 | skeletal muscle atrophy(GO:0014732) striated muscle atrophy(GO:0014891) |
| 0.1 | 1.4 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 1.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.5 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 2.3 | GO:0019374 | galactolipid metabolic process(GO:0019374) glycolipid catabolic process(GO:0019377) |
| 0.1 | 0.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.4 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 4.0 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 1.4 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.1 | 6.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.6 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.6 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 23.2 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 0.9 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 1.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 0.6 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.5 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.1 | 0.5 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 8.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 2.4 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.1 | 2.2 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.5 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.4 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.3 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.6 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 1.3 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.3 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.5 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 1.1 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 1.3 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.3 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 5.1 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 10.1 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.7 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 1.3 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 4.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.6 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.7 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 1.5 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.3 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:2001269 | interleukin-6-mediated signaling pathway(GO:0070102) positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 1.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 3.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 33.3 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 2.7 | 30.2 | GO:0045179 | apical cortex(GO:0045179) |
| 2.0 | 17.6 | GO:1990357 | terminal web(GO:1990357) |
| 1.6 | 15.8 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 1.5 | 4.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 2.2 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.6 | 2.5 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.6 | 7.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.4 | 0.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.4 | 3.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 1.5 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.3 | 3.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 1.3 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.2 | 1.9 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 4.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 31.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 13.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 2.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 39.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 4.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 13.1 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 42.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 0.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.7 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 1.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 2.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 4.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 5.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 6.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 43.4 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.2 | GO:0005767 | secondary lysosome(GO:0005767) autolysosome(GO:0044754) |
| 0.0 | 118.2 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 7.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 3.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 1.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 22.9 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 4.3 | 30.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 3.7 | 11.1 | GO:0070540 | stearic acid binding(GO:0070540) |
| 2.5 | 7.6 | GO:0035375 | zymogen binding(GO:0035375) |
| 2.3 | 9.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 2.0 | 32.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.9 | 33.3 | GO:0015250 | water channel activity(GO:0015250) |
| 1.7 | 20.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.3 | 3.9 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.2 | 10.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.1 | 32.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 1.0 | 11.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 1.0 | 3.0 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.9 | 5.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.8 | 2.5 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.7 | 21.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.6 | 3.9 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.6 | 6.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.6 | 3.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.6 | 3.0 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.6 | 13.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.5 | 3.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.5 | 4.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.5 | 8.8 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.4 | 5.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.4 | 1.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.4 | 24.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.3 | 0.8 | GO:0071820 | N-box binding(GO:0071820) |
| 0.2 | 2.4 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.2 | 22.2 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.2 | 6.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 0.9 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.2 | 2.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 1.5 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.2 | 0.8 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.2 | 1.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 0.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 0.6 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 2.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 2.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 0.5 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.1 | 6.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 14.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 15.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.4 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 17.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 2.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 8.2 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.1 | 1.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 4.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.5 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.5 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 2.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 2.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 3.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 1.0 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 4.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 18.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 3.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 4.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 1.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.1 | GO:0008823 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 37.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 17.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 13.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 5.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 8.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 11.6 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 3.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 5.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 5.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 12.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 5.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 5.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 33.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.5 | 16.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.0 | 32.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 1.0 | 34.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 1.0 | 3.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.5 | 8.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.5 | 20.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.4 | 1.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.4 | 11.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.4 | 5.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 3.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 18.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 35.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.2 | 4.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 8.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 5.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 6.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 3.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 3.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 2.1 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |