Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Hoxd12
Z-value: 1.08
Transcription factors associated with Hoxd12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd12
|
ENSMUSG00000001823.6 | Hoxd12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd12 | mm39_v1_chr2_+_74505350_74505357 | -0.18 | 1.2e-01 | Click! |
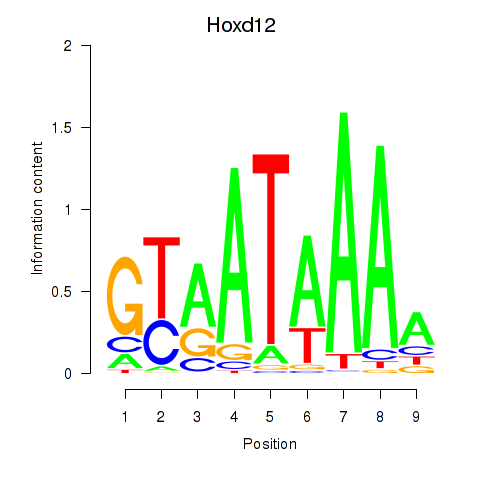
Activity profile of Hoxd12 motif
Sorted Z-values of Hoxd12 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd12
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_77255099 | 11.84 |
ENSMUST00000184138.8
ENSMUST00000184006.8 ENSMUST00000185144.8 ENSMUST00000034910.16 |
Mlip
|
muscular LMNA-interacting protein |
| chr9_-_77255069 | 11.12 |
ENSMUST00000184848.8
ENSMUST00000184415.8 |
Mlip
|
muscular LMNA-interacting protein |
| chr19_-_45800730 | 8.78 |
ENSMUST00000086993.11
|
Kcnip2
|
Kv channel-interacting protein 2 |
| chr9_-_77255171 | 8.14 |
ENSMUST00000185039.8
|
Mlip
|
muscular LMNA-interacting protein |
| chr11_-_99134885 | 7.94 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr3_-_126792056 | 7.85 |
ENSMUST00000044443.15
|
Ank2
|
ankyrin 2, brain |
| chr15_-_8740218 | 7.59 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr15_-_8739893 | 7.48 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr11_-_42070517 | 6.71 |
ENSMUST00000206105.2
ENSMUST00000153147.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr6_+_125192514 | 6.28 |
ENSMUST00000032487.14
ENSMUST00000100942.9 ENSMUST00000063588.11 |
Vamp1
|
vesicle-associated membrane protein 1 |
| chr15_+_81820954 | 6.13 |
ENSMUST00000038757.8
ENSMUST00000230633.2 |
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr14_+_26616514 | 5.89 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr10_+_86599836 | 5.82 |
ENSMUST00000218802.2
|
Gm49358
|
predicted gene, 49358 |
| chr5_-_69699965 | 5.29 |
ENSMUST00000031045.10
|
Yipf7
|
Yip1 domain family, member 7 |
| chr4_+_148085179 | 5.27 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr17_+_72088281 | 5.18 |
ENSMUST00000229874.2
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr3_-_122828592 | 4.95 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr19_-_50667079 | 4.72 |
ENSMUST00000209413.2
ENSMUST00000072685.13 ENSMUST00000164039.9 |
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr17_-_52133594 | 4.64 |
ENSMUST00000129667.8
ENSMUST00000169480.8 ENSMUST00000148559.2 |
Satb1
|
special AT-rich sequence binding protein 1 |
| chr3_+_8574420 | 4.54 |
ENSMUST00000029002.9
|
Stmn2
|
stathmin-like 2 |
| chr9_+_110948492 | 4.53 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr3_-_154760978 | 4.46 |
ENSMUST00000064076.6
|
Tnni3k
|
TNNI3 interacting kinase |
| chr3_-_117153802 | 4.37 |
ENSMUST00000197743.2
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr1_+_128069677 | 4.19 |
ENSMUST00000187023.7
|
R3hdm1
|
R3H domain containing 1 |
| chr2_+_155453103 | 4.06 |
ENSMUST00000092995.6
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr5_-_69699932 | 4.05 |
ENSMUST00000202423.2
|
Yipf7
|
Yip1 domain family, member 7 |
| chr3_+_75981577 | 3.93 |
ENSMUST00000038364.15
|
Fstl5
|
follistatin-like 5 |
| chr3_+_67799510 | 3.77 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr12_-_69838004 | 3.75 |
ENSMUST00000221646.2
|
Cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr19_-_6964988 | 3.74 |
ENSMUST00000130048.8
ENSMUST00000025914.7 |
Vegfb
|
vascular endothelial growth factor B |
| chr8_-_47805383 | 3.71 |
ENSMUST00000110367.10
|
Stox2
|
storkhead box 2 |
| chr1_-_69147185 | 3.59 |
ENSMUST00000121473.8
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chr17_+_6025861 | 3.50 |
ENSMUST00000142409.8
ENSMUST00000061091.14 |
Synj2
|
synaptojanin 2 |
| chr17_-_82045800 | 3.39 |
ENSMUST00000235015.2
ENSMUST00000163123.3 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr11_-_118246332 | 3.30 |
ENSMUST00000017610.10
|
Timp2
|
tissue inhibitor of metalloproteinase 2 |
| chr1_-_46927230 | 3.12 |
ENSMUST00000185520.2
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr3_-_141637245 | 3.02 |
ENSMUST00000106232.8
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr8_+_45960931 | 2.95 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr18_-_35348049 | 2.93 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr17_-_36207965 | 2.92 |
ENSMUST00000150056.2
ENSMUST00000156817.2 ENSMUST00000146451.8 ENSMUST00000148482.8 |
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr17_+_6026015 | 2.92 |
ENSMUST00000115790.8
|
Synj2
|
synaptojanin 2 |
| chrX_-_42363663 | 2.90 |
ENSMUST00000016294.8
|
Tenm1
|
teneurin transmembrane protein 1 |
| chr17_-_91400142 | 2.85 |
ENSMUST00000160800.9
|
Nrxn1
|
neurexin I |
| chr2_+_65760477 | 2.81 |
ENSMUST00000176109.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr2_+_65676176 | 2.79 |
ENSMUST00000053910.10
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr11_-_118246566 | 2.71 |
ENSMUST00000155707.3
|
Timp2
|
tissue inhibitor of metalloproteinase 2 |
| chr19_+_24853039 | 2.71 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr15_+_8997480 | 2.65 |
ENSMUST00000227191.3
|
Ranbp3l
|
RAN binding protein 3-like |
| chr5_+_107645626 | 2.65 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr12_+_55883101 | 2.64 |
ENSMUST00000059250.8
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr18_+_37864045 | 2.62 |
ENSMUST00000192535.2
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr3_-_26386609 | 2.60 |
ENSMUST00000193603.6
|
Nlgn1
|
neuroligin 1 |
| chr10_-_42459624 | 2.52 |
ENSMUST00000019938.11
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr8_+_45960804 | 2.50 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr7_+_30264835 | 2.50 |
ENSMUST00000043850.14
|
Igflr1
|
IGF-like family receptor 1 |
| chrX_+_35459557 | 2.48 |
ENSMUST00000115258.9
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chrX_+_35459589 | 2.45 |
ENSMUST00000048067.10
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chr12_+_103524690 | 2.40 |
ENSMUST00000187155.7
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chrX_+_35459601 | 2.40 |
ENSMUST00000115257.8
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chrM_+_3906 | 2.38 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr1_+_128069716 | 2.36 |
ENSMUST00000187557.2
|
R3hdm1
|
R3H domain containing 1 |
| chr11_+_29413734 | 2.31 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr2_-_66240408 | 2.29 |
ENSMUST00000112366.8
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr3_+_103482591 | 2.22 |
ENSMUST00000090697.11
ENSMUST00000239027.2 |
Syt6
|
synaptotagmin VI |
| chr3_+_66893280 | 2.20 |
ENSMUST00000161726.2
ENSMUST00000160504.2 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chrX_+_35459621 | 2.19 |
ENSMUST00000115256.2
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chrX_+_139357362 | 2.19 |
ENSMUST00000033809.4
|
Prps1
|
phosphoribosyl pyrophosphate synthetase 1 |
| chr14_-_36641270 | 2.15 |
ENSMUST00000182797.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr1_-_176041498 | 2.14 |
ENSMUST00000111167.2
|
Pld5
|
phospholipase D family, member 5 |
| chr14_-_67106037 | 2.10 |
ENSMUST00000022629.9
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr15_+_81629258 | 2.07 |
ENSMUST00000109554.3
ENSMUST00000230946.2 |
Zc3h7b
|
zinc finger CCCH type containing 7B |
| chr9_-_103569984 | 2.03 |
ENSMUST00000049452.15
|
Tmem108
|
transmembrane protein 108 |
| chrM_+_7758 | 2.01 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr14_-_36641470 | 2.00 |
ENSMUST00000182042.2
|
Ccser2
|
coiled-coil serine rich 2 |
| chr9_-_89620461 | 1.93 |
ENSMUST00000060700.4
ENSMUST00000185470.3 |
Ankrd34c
|
ankyrin repeat domain 34C |
| chr10_-_30531832 | 1.91 |
ENSMUST00000217138.2
ENSMUST00000217644.2 ENSMUST00000216172.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chrM_+_7779 | 1.89 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr9_+_65268304 | 1.88 |
ENSMUST00000147185.3
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr8_-_25528972 | 1.88 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4 |
| chr15_+_81821112 | 1.83 |
ENSMUST00000135663.2
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr1_-_163141278 | 1.80 |
ENSMUST00000027878.14
|
Prrx1
|
paired related homeobox 1 |
| chr4_+_136011969 | 1.77 |
ENSMUST00000144217.8
|
Zfp46
|
zinc finger protein 46 |
| chrX_-_142716200 | 1.75 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr4_+_43641262 | 1.75 |
ENSMUST00000123351.8
ENSMUST00000128549.3 |
Npr2
|
natriuretic peptide receptor 2 |
| chr10_-_30531768 | 1.72 |
ENSMUST00000092610.12
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr8_-_70355208 | 1.72 |
ENSMUST00000110167.5
|
Ndufa13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr13_-_21826688 | 1.71 |
ENSMUST00000205631.3
|
Olfr11
|
olfactory receptor 11 |
| chr10_+_45453907 | 1.70 |
ENSMUST00000037044.13
|
Hace1
|
HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 |
| chr10_+_69623262 | 1.69 |
ENSMUST00000183240.2
|
Ank3
|
ankyrin 3, epithelial |
| chr13_-_23806530 | 1.67 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr17_-_36208265 | 1.65 |
ENSMUST00000148721.8
|
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr5_-_24235646 | 1.64 |
ENSMUST00000197617.5
ENSMUST00000030849.13 |
Fam126a
|
family with sequence similarity 126, member A |
| chr3_+_66892979 | 1.63 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr10_-_35587888 | 1.56 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr10_+_119828821 | 1.54 |
ENSMUST00000105261.9
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr1_-_28819331 | 1.53 |
ENSMUST00000059937.5
|
Gm597
|
predicted gene 597 |
| chr4_+_136012019 | 1.51 |
ENSMUST00000130223.8
|
Zfp46
|
zinc finger protein 46 |
| chr2_+_73102269 | 1.47 |
ENSMUST00000090813.6
|
Sp9
|
trans-acting transcription factor 9 |
| chr15_+_36179676 | 1.47 |
ENSMUST00000171205.3
|
Spag1
|
sperm associated antigen 1 |
| chr17_+_3447465 | 1.46 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr4_-_154721288 | 1.39 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr14_-_31503869 | 1.38 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr3_+_66893031 | 1.37 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr19_+_60744385 | 1.35 |
ENSMUST00000088237.6
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr5_+_119970733 | 1.32 |
ENSMUST00000202723.4
|
Tbx5
|
T-box 5 |
| chr10_+_116013256 | 1.25 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr10_+_43400074 | 1.25 |
ENSMUST00000057649.8
ENSMUST00000216543.2 |
Gm9803
|
predicted gene 9803 |
| chr19_+_22425565 | 1.21 |
ENSMUST00000037901.14
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr5_+_20112500 | 1.12 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr13_-_43632368 | 1.09 |
ENSMUST00000222651.2
|
Ranbp9
|
RAN binding protein 9 |
| chr17_+_74111823 | 1.07 |
ENSMUST00000024860.9
|
Ehd3
|
EH-domain containing 3 |
| chr1_+_163875783 | 1.03 |
ENSMUST00000027874.6
|
Sele
|
selectin, endothelial cell |
| chr9_-_123778334 | 1.00 |
ENSMUST00000071404.5
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr5_-_74692327 | 0.98 |
ENSMUST00000072857.13
ENSMUST00000113542.9 ENSMUST00000151474.3 |
Scfd2
|
Sec1 family domain containing 2 |
| chr10_+_116013122 | 0.97 |
ENSMUST00000148731.8
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr17_+_35278011 | 0.96 |
ENSMUST00000007255.13
ENSMUST00000174493.8 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr13_+_83672389 | 0.95 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr1_-_74639723 | 0.95 |
ENSMUST00000127938.8
ENSMUST00000154874.8 |
Rnf25
|
ring finger protein 25 |
| chr10_-_25076008 | 0.95 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr5_+_143864993 | 0.95 |
ENSMUST00000172367.3
|
Gm42421
|
predicted gene, 42421 |
| chr4_+_31964082 | 0.92 |
ENSMUST00000037607.11
ENSMUST00000080933.13 ENSMUST00000108183.8 ENSMUST00000108184.3 |
Map3k7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr3_+_121838076 | 0.90 |
ENSMUST00000013995.13
|
Abca4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr12_+_33197692 | 0.90 |
ENSMUST00000077456.13
ENSMUST00000110824.9 |
Atxn7l1
|
ataxin 7-like 1 |
| chr3_+_103482547 | 0.90 |
ENSMUST00000121834.8
|
Syt6
|
synaptotagmin VI |
| chr10_-_38998272 | 0.86 |
ENSMUST00000136546.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr5_+_31212165 | 0.82 |
ENSMUST00000202795.4
ENSMUST00000201182.4 ENSMUST00000200953.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr4_-_83203388 | 0.75 |
ENSMUST00000150522.8
|
Ttc39b
|
tetratricopeptide repeat domain 39B |
| chr3_-_19217174 | 0.75 |
ENSMUST00000029125.10
|
Armc1
|
armadillo repeat containing 1 |
| chr1_+_132128701 | 0.73 |
ENSMUST00000189062.2
|
Lemd1
|
LEM domain containing 1 |
| chr4_-_82423944 | 0.69 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr7_-_102408573 | 0.67 |
ENSMUST00000210453.3
ENSMUST00000232246.3 ENSMUST00000239110.2 ENSMUST00000060187.15 ENSMUST00000168007.3 |
Olfr560
Olfr78
|
olfactory receptor 560 olfactory receptor 78 |
| chr3_+_40978804 | 0.66 |
ENSMUST00000099121.10
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr19_-_43741363 | 0.66 |
ENSMUST00000045562.6
|
Cox15
|
cytochrome c oxidase assembly protein 15 |
| chr10_+_42459772 | 0.65 |
ENSMUST00000105497.8
ENSMUST00000144806.8 |
Ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr8_+_45960855 | 0.64 |
ENSMUST00000141039.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr1_-_128030148 | 0.61 |
ENSMUST00000086614.12
|
Zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr2_-_131194754 | 0.60 |
ENSMUST00000059372.11
|
Rnf24
|
ring finger protein 24 |
| chr9_+_109641348 | 0.60 |
ENSMUST00000200555.5
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr14_+_31807760 | 0.58 |
ENSMUST00000170600.8
ENSMUST00000168986.7 ENSMUST00000169649.2 |
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr1_+_46464625 | 0.58 |
ENSMUST00000189749.7
|
Dnah7c
|
dynein, axonemal, heavy chain 7C |
| chr7_-_100232276 | 0.56 |
ENSMUST00000152876.3
ENSMUST00000150042.8 ENSMUST00000132888.9 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chrX_-_71961890 | 0.54 |
ENSMUST00000152200.2
|
Cetn2
|
centrin 2 |
| chr19_+_26725589 | 0.54 |
ENSMUST00000207812.2
ENSMUST00000175791.9 ENSMUST00000207118.2 ENSMUST00000209085.2 ENSMUST00000112637.10 ENSMUST00000207054.2 ENSMUST00000208589.2 ENSMUST00000176475.9 ENSMUST00000176698.9 ENSMUST00000207832.2 ENSMUST00000177252.9 ENSMUST00000208712.2 ENSMUST00000208186.2 ENSMUST00000208806.2 ENSMUST00000208027.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr18_-_84854841 | 0.53 |
ENSMUST00000236689.2
|
Gm17266
|
predicted gene, 17266 |
| chr6_+_90179768 | 0.53 |
ENSMUST00000078371.6
|
V1ra8
|
vomeronasal 1 receptor, A8 |
| chr6_-_38230890 | 0.51 |
ENSMUST00000117556.8
ENSMUST00000169256.5 |
D630045J12Rik
|
RIKEN cDNA D630045J12 gene |
| chr14_-_68771138 | 0.51 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr5_+_31212110 | 0.47 |
ENSMUST00000013773.12
ENSMUST00000201838.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr11_-_102210568 | 0.47 |
ENSMUST00000173870.8
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr17_+_71326542 | 0.45 |
ENSMUST00000179759.3
|
Myom1
|
myomesin 1 |
| chr17_+_35235552 | 0.44 |
ENSMUST00000007245.8
ENSMUST00000172499.2 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr4_-_82423511 | 0.43 |
ENSMUST00000050872.15
ENSMUST00000064770.9 |
Nfib
|
nuclear factor I/B |
| chr7_+_89780785 | 0.43 |
ENSMUST00000208684.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr2_-_35994072 | 0.43 |
ENSMUST00000112961.10
ENSMUST00000112966.10 |
Lhx6
|
LIM homeobox protein 6 |
| chr3_-_87676273 | 0.42 |
ENSMUST00000174776.8
|
Pear1
|
platelet endothelial aggregation receptor 1 |
| chr13_+_25127127 | 0.41 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chrX_+_105964224 | 0.39 |
ENSMUST00000060576.8
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr13_-_53627110 | 0.38 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr17_-_47169380 | 0.37 |
ENSMUST00000233455.2
|
Bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr18_-_3281752 | 0.36 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr10_-_67972401 | 0.35 |
ENSMUST00000218532.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr14_+_33774640 | 0.35 |
ENSMUST00000226211.2
|
Antxrl
|
anthrax toxin receptor-like |
| chr18_+_44249254 | 0.33 |
ENSMUST00000212114.2
|
Gm37797
|
predicted gene, 37797 |
| chr17_+_71326510 | 0.32 |
ENSMUST00000073211.13
ENSMUST00000024847.14 |
Myom1
|
myomesin 1 |
| chr3_-_154036180 | 0.29 |
ENSMUST00000177846.8
|
Lhx8
|
LIM homeobox protein 8 |
| chr1_+_59296065 | 0.28 |
ENSMUST00000160662.8
ENSMUST00000114248.3 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr5_-_25047577 | 0.28 |
ENSMUST00000030787.9
|
Rheb
|
Ras homolog enriched in brain |
| chr16_-_35589726 | 0.26 |
ENSMUST00000023554.9
|
Slc49a4
|
solute carrier family 49 member 4 |
| chr16_+_57173632 | 0.26 |
ENSMUST00000099667.3
|
Filip1l
|
filamin A interacting protein 1-like |
| chr4_-_133695264 | 0.26 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr14_-_31807552 | 0.25 |
ENSMUST00000022461.11
ENSMUST00000067955.12 ENSMUST00000124303.9 |
Dph3
|
diphthamine biosynthesis 3 |
| chr1_+_173986288 | 0.24 |
ENSMUST00000068403.4
|
Olfr420
|
olfactory receptor 420 |
| chr14_+_54413727 | 0.24 |
ENSMUST00000103701.2
|
Traj41
|
T cell receptor alpha joining 41 |
| chr14_+_53048391 | 0.24 |
ENSMUST00000103646.5
|
Trav10d
|
T cell receptor alpha variable 10D |
| chr3_-_87676239 | 0.23 |
ENSMUST00000173184.2
ENSMUST00000172621.8 ENSMUST00000174759.8 ENSMUST00000172590.8 ENSMUST00000079083.12 |
Pear1
|
platelet endothelial aggregation receptor 1 |
| chr5_-_138185686 | 0.19 |
ENSMUST00000110936.8
|
Taf6
|
TATA-box binding protein associated factor 6 |
| chr11_-_109363406 | 0.18 |
ENSMUST00000168740.3
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr15_+_39255185 | 0.18 |
ENSMUST00000228839.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr12_-_73093953 | 0.14 |
ENSMUST00000050029.8
|
Six1
|
sine oculis-related homeobox 1 |
| chr18_-_3281727 | 0.14 |
ENSMUST00000154705.8
ENSMUST00000151084.8 |
Crem
|
cAMP responsive element modulator |
| chr2_-_35994819 | 0.12 |
ENSMUST00000148852.4
|
Lhx6
|
LIM homeobox protein 6 |
| chrX_-_103244784 | 0.12 |
ENSMUST00000118314.8
|
Nexmif
|
neurite extension and migration factor |
| chr4_+_109092459 | 0.10 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chrX_-_142716085 | 0.09 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr5_-_31359559 | 0.09 |
ENSMUST00000202929.2
ENSMUST00000201231.2 ENSMUST00000114590.8 |
Zfp513
|
zinc finger protein 513 |
| chr11_-_101785181 | 0.08 |
ENSMUST00000057054.8
|
Meox1
|
mesenchyme homeobox 1 |
| chr17_+_38104420 | 0.08 |
ENSMUST00000216051.3
|
Olfr123
|
olfactory receptor 123 |
| chr10_+_89906956 | 0.08 |
ENSMUST00000183109.2
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr9_+_40092216 | 0.06 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr10_+_129153986 | 0.05 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr2_-_85478663 | 0.05 |
ENSMUST00000099920.2
|
Olfr1002
|
olfactory receptor 1002 |
| chr4_-_82423985 | 0.04 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr14_-_48900192 | 0.04 |
ENSMUST00000122009.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr2_-_18053158 | 0.03 |
ENSMUST00000066885.6
|
Skida1
|
SKI/DACH domain containing 1 |
| chr3_+_5815863 | 0.02 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 15.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.0 | 6.0 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 1.6 | 9.5 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 1.3 | 5.3 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 1.3 | 8.8 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 1.1 | 4.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.9 | 4.5 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.9 | 3.6 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.9 | 7.8 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.9 | 2.6 | GO:0048789 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.7 | 3.7 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.7 | 2.2 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.7 | 2.7 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.6 | 31.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.6 | 2.5 | GO:0021764 | amygdala development(GO:0021764) |
| 0.6 | 3.0 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.6 | 2.4 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.5 | 3.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.5 | 1.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.5 | 2.8 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.5 | 1.4 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.4 | 1.3 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.4 | 2.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.4 | 3.4 | GO:0071313 | cellular response to caffeine(GO:0071313) positive regulation of the force of heart contraction(GO:0098735) |
| 0.4 | 2.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.3 | 1.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 6.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.3 | 6.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.3 | 6.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 1.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 | 3.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 0.9 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.2 | 0.7 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.2 | 3.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 4.5 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.2 | 2.0 | GO:0097106 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) postsynaptic density organization(GO:0097106) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.2 | 4.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 1.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 0.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.2 | 1.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.2 | 1.3 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.2 | 2.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 0.9 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 6.1 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.7 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 2.9 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.4 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 0.6 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 1.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 2.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.8 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 3.9 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.4 | GO:0010982 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 6.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 4.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 1.4 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.1 | 0.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 1.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 2.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 0.7 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.4 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.5 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 8.0 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 5.7 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 5.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0072107 | myotome development(GO:0061055) regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.0 | 0.5 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 3.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.0 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 1.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 4.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.6 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 2.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 1.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.5 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.4 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 1.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.5 | 2.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.5 | 6.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.5 | 5.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.4 | 1.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.3 | 35.7 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 3.9 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.2 | 4.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 5.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 4.0 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 8.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.5 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 2.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 2.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 4.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 3.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 18.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 8.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 14.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 5.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 14.4 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 1.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.4 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 1.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 9.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 5.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 15.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.8 | 8.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.2 | 3.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.1 | 3.4 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 1.0 | 4.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.8 | 6.7 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.6 | 4.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.5 | 2.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.5 | 6.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.4 | 1.3 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.4 | 2.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.4 | 3.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.3 | 0.9 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.3 | 4.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.3 | 1.7 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.3 | 6.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 9.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.3 | 3.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 5.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.6 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.2 | 1.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.2 | 33.0 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 4.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 4.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 6.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 9.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 2.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.5 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 2.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 3.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 7.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 4.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 7.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 2.6 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 0.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.5 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 1.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 4.3 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 2.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 3.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 8.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 4.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 2.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 5.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 5.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 5.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 7.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 6.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 6.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 6.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 15.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 3.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 4.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 6.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 3.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.9 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 2.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 2.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 2.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |