Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
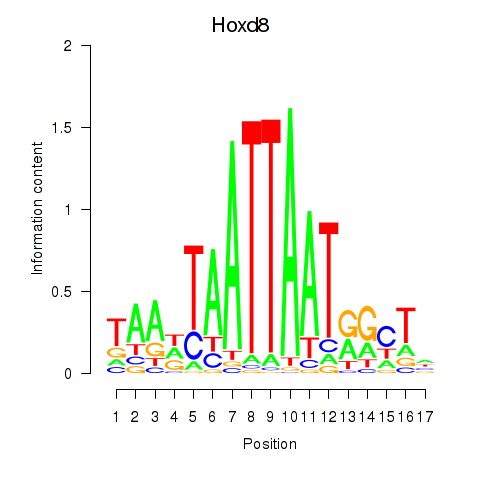
Results for Hoxd8
Z-value: 0.98
Transcription factors associated with Hoxd8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd8
|
ENSMUSG00000027102.5 | Hoxd8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd8 | mm39_v1_chr2_+_74534959_74534992 | 0.48 | 2.3e-05 | Click! |
Activity profile of Hoxd8 motif
Sorted Z-values of Hoxd8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_63036096 | 9.19 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr18_+_4165832 | 6.79 |
ENSMUST00000025076.10
|
Lyzl1
|
lysozyme-like 1 |
| chr1_+_177557380 | 6.51 |
ENSMUST00000016106.6
|
1700016C15Rik
|
RIKEN cDNA 1700016C15 gene |
| chr12_-_64521464 | 5.10 |
ENSMUST00000059833.8
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr11_-_116572116 | 5.09 |
ENSMUST00000144398.2
|
St6galnac2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
| chr3_+_18002574 | 5.01 |
ENSMUST00000029080.5
|
Cypt12
|
cysteine-rich perinuclear theca 12 |
| chr2_+_110427643 | 4.99 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr1_+_88093726 | 4.95 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr12_+_31440842 | 4.89 |
ENSMUST00000167432.8
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr4_+_52596266 | 4.86 |
ENSMUST00000029995.6
|
Toporsl
|
topoisomerase I binding, arginine/serine-rich like |
| chr18_+_4165856 | 4.71 |
ENSMUST00000120837.2
|
Lyzl1
|
lysozyme-like 1 |
| chr14_+_96118660 | 4.54 |
ENSMUST00000228913.2
ENSMUST00000045892.3 |
Spertl
|
spermatid associated like |
| chr6_+_30610973 | 4.52 |
ENSMUST00000062758.11
|
Cpa5
|
carboxypeptidase A5 |
| chr10_+_23770586 | 4.40 |
ENSMUST00000041416.8
|
Vnn1
|
vanin 1 |
| chr6_+_30611028 | 4.27 |
ENSMUST00000115138.8
|
Cpa5
|
carboxypeptidase A5 |
| chr9_-_105022272 | 4.23 |
ENSMUST00000190661.2
ENSMUST00000035180.5 |
Nudt16l2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 2 |
| chr4_+_109092610 | 4.19 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr4_+_148985584 | 4.12 |
ENSMUST00000147270.2
|
Casz1
|
castor zinc finger 1 |
| chr13_+_24023428 | 3.96 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr5_+_87148697 | 3.83 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr3_+_94280101 | 3.64 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr10_+_97318223 | 3.62 |
ENSMUST00000163448.4
|
Dcn
|
decorin |
| chr1_+_87983099 | 3.54 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr1_+_106866678 | 3.51 |
ENSMUST00000112724.3
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr1_-_186947618 | 3.44 |
ENSMUST00000110945.4
ENSMUST00000183931.8 ENSMUST00000027908.13 |
Spata17
|
spermatogenesis associated 17 |
| chr3_-_75072319 | 3.40 |
ENSMUST00000124618.2
|
Zbbx
|
zinc finger, B-box domain containing |
| chr19_-_46661321 | 3.36 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr16_-_69660606 | 3.26 |
ENSMUST00000076500.14
|
Speer2
|
spermatogenesis associated glutamate (E)-rich protein 2 |
| chr15_-_57128522 | 3.07 |
ENSMUST00000137764.2
ENSMUST00000022995.13 |
Slc22a22
|
solute carrier family 22 (organic cation transporter), member 22 |
| chr13_+_24023386 | 2.99 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr17_-_28779678 | 2.92 |
ENSMUST00000114785.3
ENSMUST00000025062.5 |
Clps
|
colipase, pancreatic |
| chr10_-_108535970 | 2.78 |
ENSMUST00000218023.2
|
Gm5136
|
predicted gene 5136 |
| chr5_-_23889591 | 2.76 |
ENSMUST00000198549.2
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr6_+_24733239 | 2.76 |
ENSMUST00000031690.6
|
Hyal6
|
hyaluronoglucosaminidase 6 |
| chr18_+_11766333 | 2.75 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr14_+_26722319 | 2.74 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr8_-_65489834 | 2.64 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr1_+_9618173 | 2.63 |
ENSMUST00000144177.8
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr7_+_45271229 | 2.61 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr4_+_109092459 | 2.61 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr19_-_46661501 | 2.58 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr8_-_65489791 | 2.56 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr13_+_49761506 | 2.53 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr4_-_96673423 | 2.47 |
ENSMUST00000107071.2
|
Gm12695
|
predicted gene 12695 |
| chr5_-_103777145 | 2.45 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr3_-_96359622 | 2.45 |
ENSMUST00000093126.11
ENSMUST00000098841.4 |
BC107364
|
cDNA sequence BC107364 |
| chr1_-_186947651 | 2.42 |
ENSMUST00000183819.8
|
Spata17
|
spermatogenesis associated 17 |
| chr1_+_87983189 | 2.40 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr11_+_117700479 | 2.40 |
ENSMUST00000026649.14
ENSMUST00000177131.8 ENSMUST00000120928.2 ENSMUST00000175737.2 ENSMUST00000132298.2 |
Syngr2
Gm20708
|
synaptogyrin 2 predicted gene 20708 |
| chrX_-_125723491 | 2.35 |
ENSMUST00000081074.5
|
4932411N23Rik
|
RIKEN cDNA 4932411N23 gene |
| chr7_+_134870237 | 2.35 |
ENSMUST00000210697.2
ENSMUST00000097983.5 |
Nps
|
neuropeptide S |
| chr18_-_43610829 | 2.28 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr7_-_12829100 | 2.25 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr1_-_134477400 | 2.21 |
ENSMUST00000172898.2
|
Mgat4e
|
MGAT4 family, member E |
| chr5_+_29400981 | 2.14 |
ENSMUST00000160888.8
ENSMUST00000159272.8 ENSMUST00000001247.12 ENSMUST00000161398.8 ENSMUST00000160246.8 |
Rnf32
|
ring finger protein 32 |
| chr10_-_103072141 | 2.14 |
ENSMUST00000123364.2
ENSMUST00000166240.8 ENSMUST00000020043.12 |
Lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr3_-_30194559 | 2.10 |
ENSMUST00000108271.10
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr7_-_44711771 | 2.08 |
ENSMUST00000210101.2
ENSMUST00000209219.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr9_+_80072361 | 2.07 |
ENSMUST00000184480.8
|
Myo6
|
myosin VI |
| chr5_-_86780277 | 2.05 |
ENSMUST00000116553.9
|
Tmprss11f
|
transmembrane protease, serine 11f |
| chr8_-_22396428 | 2.03 |
ENSMUST00000051965.5
|
Defb11
|
defensin beta 11 |
| chr11_-_59466995 | 1.97 |
ENSMUST00000215339.2
ENSMUST00000214351.2 |
Olfr222
|
olfactory receptor 222 |
| chr10_-_127843377 | 1.96 |
ENSMUST00000219447.2
ENSMUST00000219780.2 ENSMUST00000219707.2 ENSMUST00000219953.2 ENSMUST00000219183.2 |
Hsd17b6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr1_+_180878797 | 1.90 |
ENSMUST00000036819.7
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr5_+_65505657 | 1.89 |
ENSMUST00000031096.11
|
Klb
|
klotho beta |
| chr9_+_80072274 | 1.82 |
ENSMUST00000035889.15
ENSMUST00000113268.8 |
Myo6
|
myosin VI |
| chr7_+_140343652 | 1.75 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr9_-_15212849 | 1.75 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chrX_-_21041892 | 1.74 |
ENSMUST00000096511.5
|
Gm21876
|
predicted gene, 21876 |
| chr8_+_46463633 | 1.74 |
ENSMUST00000110381.9
|
Lrp2bp
|
Lrp2 binding protein |
| chr9_-_55419442 | 1.68 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr2_+_89757653 | 1.59 |
ENSMUST00000213720.3
ENSMUST00000102609.3 |
Olfr1258
|
olfactory receptor 1258 |
| chr7_-_80055168 | 1.51 |
ENSMUST00000107362.10
ENSMUST00000135306.3 |
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr9_+_21634779 | 1.49 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr10_+_116881246 | 1.48 |
ENSMUST00000073834.5
|
Lrrc10
|
leucine rich repeat containing 10 |
| chrY_+_65387652 | 1.46 |
ENSMUST00000178198.2
|
Gm20736
|
predicted gene, 20736 |
| chr1_-_126758369 | 1.44 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr7_+_30193047 | 1.42 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chrX_-_105884178 | 1.42 |
ENSMUST00000062010.10
|
Rtl3
|
retrotransposon Gag like 3 |
| chr4_-_132125510 | 1.41 |
ENSMUST00000136711.2
ENSMUST00000084249.11 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr4_+_109092829 | 1.39 |
ENSMUST00000030285.8
|
Calr4
|
calreticulin 4 |
| chr16_+_14407073 | 1.35 |
ENSMUST00000100165.4
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chrY_+_79332266 | 1.34 |
ENSMUST00000178063.2
|
Gm20916
|
predicted gene, 20916 |
| chrY_+_52778041 | 1.34 |
ENSMUST00000178673.2
|
Gm21258
|
predicted gene, 21258 |
| chr1_+_88234454 | 1.29 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr2_+_110551976 | 1.28 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
| chr15_-_96929086 | 1.28 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chrY_-_35875442 | 1.27 |
ENSMUST00000180332.2
|
Gm20896
|
predicted gene, 20896 |
| chrY_+_84109980 | 1.27 |
ENSMUST00000177775.2
|
Gm21095
|
predicted gene, 21095 |
| chr7_+_130633776 | 1.26 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chrY_+_55211732 | 1.26 |
ENSMUST00000180249.2
|
Gm20931
|
predicted gene, 20931 |
| chr10_-_126866682 | 1.23 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr11_-_99265721 | 1.22 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chrY_-_40270796 | 1.20 |
ENSMUST00000177713.2
|
Gm21865
|
predicted gene, 21865 |
| chr2_+_152528955 | 1.19 |
ENSMUST00000062148.9
|
Mcts2
|
malignant T cell amplified sequence 2 |
| chr3_+_93107517 | 1.19 |
ENSMUST00000098884.4
|
Flg2
|
filaggrin family member 2 |
| chr2_+_178072324 | 1.19 |
ENSMUST00000108912.9
ENSMUST00000042092.9 |
Cdh26
|
cadherin-like 26 |
| chr15_+_99192968 | 1.15 |
ENSMUST00000128352.8
ENSMUST00000145482.8 |
Prpf40b
|
pre-mRNA processing factor 40B |
| chr8_+_22329942 | 1.14 |
ENSMUST00000006745.4
|
Defb2
|
defensin beta 2 |
| chr9_+_38240356 | 1.13 |
ENSMUST00000214155.2
ENSMUST00000212354.3 |
Olfr25
|
olfactory receptor 25 |
| chr16_-_75382905 | 1.13 |
ENSMUST00000062721.11
|
Lipi
|
lipase, member I |
| chr14_+_47536075 | 1.12 |
ENSMUST00000227554.2
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr4_+_145397238 | 1.11 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chrY_+_55729767 | 1.10 |
ENSMUST00000177834.2
|
Gm21858
|
predicted gene, 21858 |
| chr1_+_107288928 | 1.09 |
ENSMUST00000191425.7
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr2_+_128704867 | 1.08 |
ENSMUST00000110324.8
|
Fbln7
|
fibulin 7 |
| chr9_+_54446268 | 1.08 |
ENSMUST00000060242.12
|
Sh2d7
|
SH2 domain containing 7 |
| chr14_+_55797443 | 1.02 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr3_-_144555062 | 1.02 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr10_+_34265969 | 0.99 |
ENSMUST00000105511.2
|
Col10a1
|
collagen, type X, alpha 1 |
| chrY_+_80146479 | 0.99 |
ENSMUST00000179811.2
|
Gm21760
|
predicted gene, 21760 |
| chr19_-_12868022 | 0.93 |
ENSMUST00000081236.3
|
Olfr1446
|
olfactory receptor 1446 |
| chr1_+_58841808 | 0.91 |
ENSMUST00000190213.2
|
Casp8
|
caspase 8 |
| chr2_-_151510453 | 0.90 |
ENSMUST00000180195.8
ENSMUST00000096439.4 |
Rad21l
|
RAD21-like (S. pombe) |
| chr2_-_88854443 | 0.90 |
ENSMUST00000099799.5
|
Olfr1217
|
olfactory receptor 1217 |
| chr17_+_46807637 | 0.89 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr6_-_141719536 | 0.89 |
ENSMUST00000148411.2
|
Gm5724
|
predicted gene 5724 |
| chr2_-_163314480 | 0.88 |
ENSMUST00000109418.2
|
Fitm2
|
fat storage-inducing transmembrane protein 2 |
| chr2_+_83554868 | 0.84 |
ENSMUST00000111740.9
|
Itgav
|
integrin alpha V |
| chr9_+_72714156 | 0.83 |
ENSMUST00000055535.9
|
Prtg
|
protogenin |
| chr14_+_55797468 | 0.82 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr10_+_129298547 | 0.80 |
ENSMUST00000077836.3
|
Olfr787
|
olfactory receptor 787 |
| chr6_-_129077867 | 0.80 |
ENSMUST00000032258.8
|
Clec2e
|
C-type lectin domain family 2, member e |
| chr6_-_30936013 | 0.79 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr17_+_74796473 | 0.79 |
ENSMUST00000024873.7
|
Yipf4
|
Yip1 domain family, member 4 |
| chr10_+_129691236 | 0.78 |
ENSMUST00000204622.3
|
Olfr813
|
olfactory receptor 813 |
| chr10_-_75946790 | 0.78 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr1_-_126758520 | 0.77 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr3_-_96104886 | 0.76 |
ENSMUST00000016087.4
|
Bola1
|
bolA-like 1 (E. coli) |
| chr5_+_68189116 | 0.75 |
ENSMUST00000094715.8
|
Grxcr1
|
glutaredoxin, cysteine rich 1 |
| chr16_-_19443851 | 0.73 |
ENSMUST00000079891.4
|
Olfr171
|
olfactory receptor 171 |
| chr2_+_85868891 | 0.73 |
ENSMUST00000218397.2
|
Olfr1033
|
olfactory receptor 1033 |
| chr2_-_85632888 | 0.71 |
ENSMUST00000217410.3
ENSMUST00000216425.3 |
Olfr1016
|
olfactory receptor 1016 |
| chr6_+_41331039 | 0.70 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr10_-_126866658 | 0.69 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr16_-_19425453 | 0.67 |
ENSMUST00000078603.3
ENSMUST00000218837.2 |
Olfr170
|
olfactory receptor 170 |
| chr11_-_99979052 | 0.66 |
ENSMUST00000107419.2
|
Krt32
|
keratin 32 |
| chr2_+_110551927 | 0.66 |
ENSMUST00000111017.9
|
Muc15
|
mucin 15 |
| chr14_+_26616514 | 0.65 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr6_+_121983720 | 0.64 |
ENSMUST00000081777.8
|
Mug2
|
murinoglobulin 2 |
| chrY_-_35085749 | 0.63 |
ENSMUST00000180170.2
|
Gm20855
|
predicted gene, 20855 |
| chr8_-_117809188 | 0.62 |
ENSMUST00000109093.9
ENSMUST00000098375.6 |
Pkd1l2
|
polycystic kidney disease 1 like 2 |
| chr2_+_152511381 | 0.59 |
ENSMUST00000125366.8
ENSMUST00000109825.8 ENSMUST00000089059.9 ENSMUST00000079247.4 |
H13
|
histocompatibility 13 |
| chr1_+_178233640 | 0.58 |
ENSMUST00000027775.9
|
Efcab2
|
EF-hand calcium binding domain 2 |
| chr1_-_144427302 | 0.57 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr2_+_87725306 | 0.57 |
ENSMUST00000217436.2
|
Olfr1153
|
olfactory receptor 1153 |
| chr2_+_83554741 | 0.57 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr3_-_69767208 | 0.56 |
ENSMUST00000171529.4
ENSMUST00000051239.13 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr3_+_142300601 | 0.56 |
ENSMUST00000029936.5
|
Gbp2b
|
guanylate binding protein 2b |
| chr17_-_56343625 | 0.56 |
ENSMUST00000003268.11
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr14_+_69585036 | 0.54 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr8_-_19114826 | 0.51 |
ENSMUST00000075504.3
|
Defb39
|
defensin beta 39 |
| chr11_-_87783073 | 0.49 |
ENSMUST00000213672.2
ENSMUST00000213928.2 |
Olfr462
|
olfactory receptor 462 |
| chr2_+_110551685 | 0.48 |
ENSMUST00000111016.9
|
Muc15
|
mucin 15 |
| chr9_-_40015750 | 0.48 |
ENSMUST00000213858.2
|
Olfr984
|
olfactory receptor 984 |
| chr13_+_65298054 | 0.47 |
ENSMUST00000214214.2
|
Olfr466
|
olfactory receptor 466 |
| chr5_+_88523967 | 0.45 |
ENSMUST00000073363.2
|
Amtn
|
amelotin |
| chr15_-_101422054 | 0.42 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chr18_-_70409277 | 0.39 |
ENSMUST00000239144.2
|
Gm36255
|
predicted gene, 36255 |
| chr19_-_13828056 | 0.39 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr2_+_89642395 | 0.38 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr17_+_79919267 | 0.38 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr3_-_95778679 | 0.36 |
ENSMUST00000142437.2
ENSMUST00000067298.5 |
Mrps21
|
mitochondrial ribosomal protein S21 |
| chr15_-_35155896 | 0.35 |
ENSMUST00000067033.8
ENSMUST00000018476.14 |
Stk3
|
serine/threonine kinase 3 |
| chr11_-_74238498 | 0.35 |
ENSMUST00000080365.6
|
Olfr411
|
olfactory receptor 411 |
| chr2_-_88909981 | 0.35 |
ENSMUST00000213724.2
|
Olfr1219
|
olfactory receptor 1219 |
| chr1_-_14380032 | 0.34 |
ENSMUST00000187790.2
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr1_-_166509404 | 0.33 |
ENSMUST00000148677.2
ENSMUST00000027843.11 |
Fmo9
|
flavin containing monooxygenase 9 |
| chr2_-_73410632 | 0.32 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr7_+_103628383 | 0.30 |
ENSMUST00000098185.2
|
Olfr635
|
olfactory receptor 635 |
| chr3_+_20011405 | 0.30 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr4_+_115099237 | 0.30 |
ENSMUST00000118278.2
|
Cyp4a29
|
cytochrome P450, family 4, subfamily a, polypeptide 29 |
| chr9_+_111011327 | 0.28 |
ENSMUST00000216430.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr7_-_103191732 | 0.27 |
ENSMUST00000215663.2
|
Olfr612
|
olfactory receptor 612 |
| chr4_+_118521590 | 0.24 |
ENSMUST00000217013.2
|
Olfr62
|
olfactory receptor 62 |
| chr14_-_50476340 | 0.23 |
ENSMUST00000059565.2
|
Olfr731
|
olfactory receptor 731 |
| chr2_+_83554770 | 0.22 |
ENSMUST00000141725.3
|
Itgav
|
integrin alpha V |
| chr6_-_3399451 | 0.22 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr10_+_129297731 | 0.21 |
ENSMUST00000213329.2
|
Olfr787
|
olfactory receptor 787 |
| chr2_+_87686206 | 0.21 |
ENSMUST00000217376.2
|
Olfr1151
|
olfactory receptor 1151 |
| chr19_-_11833365 | 0.21 |
ENSMUST00000079875.4
|
Olfr1418
|
olfactory receptor 1418 |
| chr8_+_34089597 | 0.20 |
ENSMUST00000009774.11
|
Ppp2cb
|
protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform |
| chr13_+_22474090 | 0.19 |
ENSMUST00000228382.2
ENSMUST00000228557.2 ENSMUST00000226245.2 ENSMUST00000227516.2 |
Vmn1r196
|
vomeronasal 1 receptor 196 |
| chr17_+_37978659 | 0.19 |
ENSMUST00000216551.2
|
Olfr118
|
olfactory receptor 118 |
| chr17_-_56343531 | 0.19 |
ENSMUST00000233803.2
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr5_+_48529671 | 0.19 |
ENSMUST00000196950.5
ENSMUST00000030968.7 |
5730480H06Rik
|
RIKEN cDNA 5730480H06 gene |
| chr7_+_102526329 | 0.17 |
ENSMUST00000098216.2
|
Olfr568
|
olfactory receptor 568 |
| chr2_+_88479038 | 0.17 |
ENSMUST00000120518.4
|
Olfr1192-ps1
|
olfactory receptor 1192, pseudogene 1 |
| chr2_+_85677374 | 0.17 |
ENSMUST00000215347.3
|
Olfr1020
|
olfactory receptor 1020 |
| chr1_-_138776315 | 0.17 |
ENSMUST00000112030.9
|
Lhx9
|
LIM homeobox protein 9 |
| chr10_-_129565161 | 0.15 |
ENSMUST00000204717.3
ENSMUST00000216794.2 ENSMUST00000217219.2 |
Olfr805
|
olfactory receptor 805 |
| chr17_-_8366536 | 0.15 |
ENSMUST00000231927.2
|
Rnaset2a
|
ribonuclease T2A |
| chr6_+_65502344 | 0.15 |
ENSMUST00000212375.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr14_+_47535717 | 0.14 |
ENSMUST00000166743.9
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr17_+_37977879 | 0.14 |
ENSMUST00000215811.2
|
Olfr118
|
olfactory receptor 118 |
| chr2_-_111820618 | 0.13 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr16_+_33614715 | 0.10 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr9_+_111011388 | 0.10 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr9_-_117843228 | 0.10 |
ENSMUST00000187803.3
|
Zcwpw2
|
zinc finger, CW type with PWWP domain 2 |
| chr5_-_38649291 | 0.09 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.2 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 1.4 | 4.1 | GO:0061076 | camera-type eye photoreceptor cell fate commitment(GO:0060220) negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 0.9 | 10.9 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.7 | 5.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.7 | 11.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.6 | 3.1 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.5 | 1.6 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.5 | 1.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.5 | 1.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.5 | 6.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.5 | 3.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.4 | 2.4 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.4 | 1.5 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.4 | 1.5 | GO:0090118 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.4 | 1.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.3 | 3.6 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.3 | 4.9 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.3 | 2.8 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.3 | 5.9 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.2 | 2.7 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 1.9 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 4.4 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 | 1.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 0.6 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 | 3.9 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 0.4 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.1 | 2.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 1.3 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.9 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.9 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.9 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 2.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.8 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 0.4 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.6 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 0.9 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 1.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.7 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.1 | 2.9 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.8 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 5.2 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 2.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.8 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.3 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 1.9 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.6 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 1.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.2 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 1.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 7.4 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 4.0 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 1.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.6 | GO:0006818 | hydrogen transport(GO:0006818) |
| 0.0 | 3.7 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 3.7 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 1.5 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.4 | 1.7 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.4 | 1.6 | GO:0034686 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.3 | 2.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 0.9 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.2 | 5.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 3.6 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.2 | 0.9 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 4.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.5 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 2.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 3.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.6 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 7.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 1.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.6 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 2.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 6.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 3.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 2.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 5.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.9 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.6 | 11.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.6 | 3.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.6 | 9.2 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.6 | 5.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.6 | 4.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.5 | 1.9 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.4 | 2.6 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.4 | 1.3 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.4 | 14.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 4.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 8.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 5.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.3 | 2.0 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.3 | 2.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 2.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 0.9 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 3.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.2 | 1.4 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.2 | 2.8 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 2.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 0.6 | GO:0019002 | GMP binding(GO:0019002) |
| 0.2 | 1.5 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.2 | 1.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 5.1 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 1.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 1.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.5 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 1.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 3.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 2.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 5.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.9 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 2.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 9.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 4.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 2.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 4.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 1.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 2.2 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.8 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 3.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.3 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.5 | 5.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.4 | 5.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 7.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 3.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 2.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 3.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 9.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 2.9 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 2.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.9 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.7 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 2.9 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |