Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
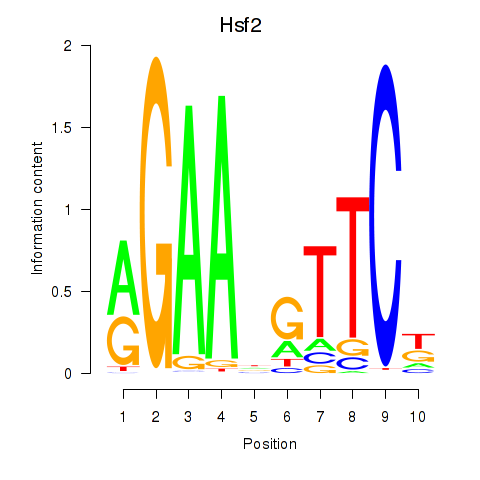
Results for Hsf2
Z-value: 1.01
Transcription factors associated with Hsf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf2
|
ENSMUSG00000019878.9 | Hsf2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf2 | mm39_v1_chr10_+_57362512_57362523 | 0.27 | 2.4e-02 | Click! |
Activity profile of Hsf2 motif
Sorted Z-values of Hsf2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_140343652 | 4.66 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr9_-_110818679 | 4.31 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chr5_+_92750898 | 4.22 |
ENSMUST00000200941.2
ENSMUST00000050952.4 |
Stbd1
|
starch binding domain 1 |
| chr5_+_135916764 | 4.07 |
ENSMUST00000005077.7
|
Hspb1
|
heat shock protein 1 |
| chrX_+_138701544 | 3.93 |
ENSMUST00000054889.4
|
Cldn2
|
claudin 2 |
| chr5_+_17779273 | 3.93 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr5_+_135916847 | 3.86 |
ENSMUST00000111155.2
|
Hspb1
|
heat shock protein 1 |
| chr3_-_107851021 | 3.54 |
ENSMUST00000106684.8
ENSMUST00000106685.9 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr5_-_87716882 | 3.40 |
ENSMUST00000113314.3
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr3_-_107925122 | 3.37 |
ENSMUST00000126593.3
|
Gstm1
|
glutathione S-transferase, mu 1 |
| chr5_+_17779721 | 3.17 |
ENSMUST00000169603.2
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr6_-_3988900 | 3.12 |
ENSMUST00000183682.3
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr15_+_100251030 | 3.12 |
ENSMUST00000075675.7
ENSMUST00000088142.6 ENSMUST00000176287.2 |
Methig1
Mettl7a2
|
methyltransferase hypoxia inducible domain containing 1 methyltransferase like 7A2 |
| chr6_+_72575458 | 3.08 |
ENSMUST00000070597.13
ENSMUST00000176364.8 ENSMUST00000176168.3 |
Retsat
|
retinol saturase (all trans retinol 13,14 reductase) |
| chr19_-_44017637 | 2.96 |
ENSMUST00000026211.10
ENSMUST00000211830.2 |
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr15_+_100202642 | 2.89 |
ENSMUST00000067752.5
ENSMUST00000229588.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr17_+_27248233 | 2.87 |
ENSMUST00000053683.7
ENSMUST00000236222.2 |
Ggnbp1
|
gametogenetin binding protein 1 |
| chr3_-_107925159 | 2.86 |
ENSMUST00000004140.11
|
Gstm1
|
glutathione S-transferase, mu 1 |
| chr11_+_114741948 | 2.83 |
ENSMUST00000133245.2
ENSMUST00000122967.3 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr6_-_3988835 | 2.80 |
ENSMUST00000203257.2
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr11_+_114742331 | 2.75 |
ENSMUST00000177952.8
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr14_+_51328534 | 2.75 |
ENSMUST00000022428.13
ENSMUST00000171688.9 |
Rnase4
Ang
|
ribonuclease, RNase A family 4 angiogenin, ribonuclease, RNase A family, 5 |
| chr11_+_101137786 | 2.75 |
ENSMUST00000107282.4
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr1_-_66974694 | 2.68 |
ENSMUST00000186202.7
|
Myl1
|
myosin, light polypeptide 1 |
| chr7_+_51160754 | 2.67 |
ENSMUST00000043944.6
ENSMUST00000207044.2 |
Ano5
|
anoctamin 5 |
| chr2_+_164404499 | 2.56 |
ENSMUST00000017867.10
ENSMUST00000109344.9 ENSMUST00000109345.9 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr19_+_4036562 | 2.55 |
ENSMUST00000236224.2
ENSMUST00000236510.2 ENSMUST00000237910.2 ENSMUST00000235612.2 ENSMUST00000054030.8 |
Acy3
|
aspartoacylase (aminoacylase) 3 |
| chr1_-_66974492 | 2.54 |
ENSMUST00000120415.8
ENSMUST00000119429.8 |
Myl1
|
myosin, light polypeptide 1 |
| chr5_-_50216249 | 2.54 |
ENSMUST00000030971.7
|
Adgra3
|
adhesion G protein-coupled receptor A3 |
| chrX_+_21350783 | 2.51 |
ENSMUST00000089188.9
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr4_-_86587728 | 2.50 |
ENSMUST00000149700.8
|
Plin2
|
perilipin 2 |
| chr7_-_79392763 | 2.49 |
ENSMUST00000032761.8
|
Pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr2_+_158148413 | 2.49 |
ENSMUST00000109491.8
ENSMUST00000016168.9 |
Lbp
|
lipopolysaccharide binding protein |
| chr10_+_40505985 | 2.46 |
ENSMUST00000019977.8
ENSMUST00000214102.2 ENSMUST00000213503.2 ENSMUST00000213442.2 ENSMUST00000216830.2 |
Ddo
|
D-aspartate oxidase |
| chr5_-_5564730 | 2.42 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr2_-_10135449 | 2.40 |
ENSMUST00000042290.14
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr7_+_121464254 | 2.39 |
ENSMUST00000033161.7
|
Scnn1b
|
sodium channel, nonvoltage-gated 1 beta |
| chr17_-_74354844 | 2.37 |
ENSMUST00000043458.9
|
Srd5a2
|
steroid 5 alpha-reductase 2 |
| chr15_+_10314173 | 2.33 |
ENSMUST00000127467.3
|
Prlr
|
prolactin receptor |
| chr5_+_92479687 | 2.33 |
ENSMUST00000125462.8
ENSMUST00000113083.9 ENSMUST00000121096.8 |
Art3
|
ADP-ribosyltransferase 3 |
| chr3_+_85948030 | 2.29 |
ENSMUST00000238545.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr4_+_150939521 | 2.28 |
ENSMUST00000030811.2
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr9_-_76953070 | 2.27 |
ENSMUST00000034911.7
|
Tinag
|
tubulointerstitial nephritis antigen |
| chr12_-_17374704 | 2.26 |
ENSMUST00000020884.16
ENSMUST00000095820.12 ENSMUST00000221129.2 ENSMUST00000127185.8 |
Atp6v1c2
|
ATPase, H+ transporting, lysosomal V1 subunit C2 |
| chr7_+_112278520 | 2.25 |
ENSMUST00000084705.13
ENSMUST00000239442.2 ENSMUST00000239404.2 ENSMUST00000059768.18 |
Tead1
|
TEA domain family member 1 |
| chr11_+_49500090 | 2.24 |
ENSMUST00000020617.3
|
Flt4
|
FMS-like tyrosine kinase 4 |
| chr1_+_88015524 | 2.23 |
ENSMUST00000113139.2
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr16_+_17712061 | 2.23 |
ENSMUST00000046937.4
|
Tssk1
|
testis-specific serine kinase 1 |
| chr7_+_79392888 | 2.22 |
ENSMUST00000035622.8
|
Wdr93
|
WD repeat domain 93 |
| chr7_+_51160855 | 2.21 |
ENSMUST00000207717.2
|
Ano5
|
anoctamin 5 |
| chr17_+_26094466 | 2.20 |
ENSMUST00000169308.8
ENSMUST00000169085.8 |
Mettl26
|
methyltransferase like 26 |
| chr5_+_104582978 | 2.19 |
ENSMUST00000086833.13
ENSMUST00000031243.15 ENSMUST00000112748.8 ENSMUST00000112746.8 ENSMUST00000145084.8 ENSMUST00000132457.8 |
Spp1
|
secreted phosphoprotein 1 |
| chr6_+_121277693 | 2.07 |
ENSMUST00000142419.2
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chrX_+_162873183 | 2.04 |
ENSMUST00000015545.10
|
Cltrn
|
collectrin, amino acid transport regulator |
| chr3_-_107850707 | 2.03 |
ENSMUST00000106681.3
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr7_-_101582987 | 2.02 |
ENSMUST00000106964.8
ENSMUST00000106963.2 ENSMUST00000078448.11 ENSMUST00000106966.8 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr10_-_126906123 | 2.02 |
ENSMUST00000060991.6
|
Tspan31
|
tetraspanin 31 |
| chr14_-_22039543 | 2.00 |
ENSMUST00000043409.9
|
Zfp503
|
zinc finger protein 503 |
| chr10_+_4660119 | 1.99 |
ENSMUST00000105588.8
ENSMUST00000105589.2 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr18_+_20798337 | 1.99 |
ENSMUST00000075312.5
|
Ttr
|
transthyretin |
| chr5_+_92479677 | 1.97 |
ENSMUST00000154245.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr3_-_107850666 | 1.95 |
ENSMUST00000106683.8
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr12_+_75355082 | 1.95 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr18_-_10706701 | 1.95 |
ENSMUST00000002549.9
ENSMUST00000117726.9 ENSMUST00000117828.9 |
Abhd3
|
abhydrolase domain containing 3 |
| chr7_+_127864847 | 1.94 |
ENSMUST00000118169.8
ENSMUST00000206909.2 ENSMUST00000142841.8 |
Slc5a2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr6_-_85690187 | 1.93 |
ENSMUST00000160534.2
|
Nat8f7
|
N-acetyltransferase 8 (GCN5-related) family member 7 |
| chr19_-_8382424 | 1.91 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chrX_-_74886623 | 1.90 |
ENSMUST00000114057.8
|
Pls3
|
plastin 3 (T-isoform) |
| chr5_-_5564873 | 1.90 |
ENSMUST00000060947.14
|
Cldn12
|
claudin 12 |
| chr4_-_82423985 | 1.89 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr19_+_20470056 | 1.86 |
ENSMUST00000225337.3
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr11_+_101137231 | 1.86 |
ENSMUST00000122006.8
ENSMUST00000151830.2 |
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr4_+_62883796 | 1.86 |
ENSMUST00000030043.13
ENSMUST00000107415.8 ENSMUST00000064814.6 |
Zfp618
|
zinc finger protein 618 |
| chr19_-_7779943 | 1.85 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr7_-_79382503 | 1.84 |
ENSMUST00000032762.14
ENSMUST00000205915.2 |
Plin1
|
perilipin 1 |
| chr3_+_85947806 | 1.83 |
ENSMUST00000238222.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr15_-_102112657 | 1.80 |
ENSMUST00000231030.2
ENSMUST00000230687.2 ENSMUST00000229514.2 ENSMUST00000229345.2 |
Csad
|
cysteine sulfinic acid decarboxylase |
| chr19_+_20470114 | 1.79 |
ENSMUST00000225313.2
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr15_+_78726824 | 1.77 |
ENSMUST00000059619.3
|
Cdc42ep1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr14_+_19801333 | 1.76 |
ENSMUST00000022340.5
|
Nid2
|
nidogen 2 |
| chr2_+_91086489 | 1.74 |
ENSMUST00000154959.8
ENSMUST00000059566.11 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr1_+_171052623 | 1.74 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr9_+_65536892 | 1.73 |
ENSMUST00000169003.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr15_+_100232810 | 1.72 |
ENSMUST00000075420.6
|
Mettl7a3
|
methyltransferase like 7A3 |
| chr7_+_112278534 | 1.71 |
ENSMUST00000106638.10
|
Tead1
|
TEA domain family member 1 |
| chr17_+_79934096 | 1.71 |
ENSMUST00000224618.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr7_-_30814652 | 1.69 |
ENSMUST00000168884.8
ENSMUST00000108102.9 |
Hpn
|
hepsin |
| chr3_+_151143557 | 1.69 |
ENSMUST00000196970.3
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr18_-_53551127 | 1.67 |
ENSMUST00000025419.9
|
Ppic
|
peptidylprolyl isomerase C |
| chr5_+_32768515 | 1.67 |
ENSMUST00000202543.4
ENSMUST00000072311.13 |
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr19_-_30152814 | 1.66 |
ENSMUST00000025778.9
|
Gldc
|
glycine decarboxylase |
| chr19_-_4889314 | 1.65 |
ENSMUST00000235245.2
ENSMUST00000037246.7 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr7_-_89176294 | 1.65 |
ENSMUST00000207932.2
|
Prss23
|
protease, serine 23 |
| chr6_-_72212547 | 1.64 |
ENSMUST00000042646.8
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr11_+_101258368 | 1.63 |
ENSMUST00000019469.3
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr18_+_36475573 | 1.63 |
ENSMUST00000139727.3
ENSMUST00000237375.2 ENSMUST00000235403.2 ENSMUST00000236593.2 ENSMUST00000236374.2 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr19_-_7780025 | 1.62 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr14_+_70077841 | 1.62 |
ENSMUST00000022678.5
|
Pebp4
|
phosphatidylethanolamine binding protein 4 |
| chr3_+_20039775 | 1.62 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin |
| chr3_-_57202301 | 1.62 |
ENSMUST00000171384.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr6_-_83513184 | 1.61 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr18_+_11052458 | 1.60 |
ENSMUST00000047762.10
|
Gata6
|
GATA binding protein 6 |
| chr7_+_99827886 | 1.60 |
ENSMUST00000207358.2
ENSMUST00000207995.2 ENSMUST00000049333.13 ENSMUST00000170954.10 ENSMUST00000179842.3 ENSMUST00000208260.2 |
Kcne3
|
potassium voltage-gated channel, Isk-related subfamily, gene 3 |
| chr6_+_29433247 | 1.60 |
ENSMUST00000101617.9
ENSMUST00000065090.8 |
Flnc
|
filamin C, gamma |
| chr5_-_87288177 | 1.59 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr11_+_82802079 | 1.58 |
ENSMUST00000018989.14
ENSMUST00000164945.3 |
Unc45b
|
unc-45 myosin chaperone B |
| chr2_+_58457370 | 1.58 |
ENSMUST00000071543.12
|
Upp2
|
uridine phosphorylase 2 |
| chr19_-_4889284 | 1.57 |
ENSMUST00000236451.2
ENSMUST00000236178.2 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr5_-_66309244 | 1.57 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr19_-_46661501 | 1.57 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr4_-_43656437 | 1.55 |
ENSMUST00000030192.5
|
Hint2
|
histidine triad nucleotide binding protein 2 |
| chrX_+_55825033 | 1.54 |
ENSMUST00000114772.9
ENSMUST00000114768.10 ENSMUST00000155882.8 |
Fhl1
|
four and a half LIM domains 1 |
| chr9_+_44309727 | 1.52 |
ENSMUST00000213268.2
|
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr3_+_40978804 | 1.51 |
ENSMUST00000099121.10
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr4_-_107780716 | 1.49 |
ENSMUST00000106719.8
ENSMUST00000106720.9 ENSMUST00000131644.2 ENSMUST00000030345.15 |
Cpt2
|
carnitine palmitoyltransferase 2 |
| chr8_-_105350898 | 1.48 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr7_-_44465043 | 1.46 |
ENSMUST00000107893.9
|
Atf5
|
activating transcription factor 5 |
| chr5_+_32768591 | 1.46 |
ENSMUST00000168707.6
|
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr8_-_3517617 | 1.46 |
ENSMUST00000111081.10
ENSMUST00000004686.13 |
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr4_+_95467653 | 1.45 |
ENSMUST00000043335.11
|
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr2_-_64806106 | 1.44 |
ENSMUST00000156765.2
|
Grb14
|
growth factor receptor bound protein 14 |
| chr3_+_96737385 | 1.42 |
ENSMUST00000058865.14
|
Pdzk1
|
PDZ domain containing 1 |
| chr6_-_92920466 | 1.42 |
ENSMUST00000113438.8
|
Adamts9
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 9 |
| chr5_-_5714196 | 1.42 |
ENSMUST00000196165.5
ENSMUST00000061008.10 ENSMUST00000135252.3 ENSMUST00000054865.13 |
Cfap69
|
cilia and flagella associated protein 69 |
| chr6_+_124470053 | 1.40 |
ENSMUST00000049124.10
|
C1rl
|
complement component 1, r subcomponent-like |
| chr4_-_141553306 | 1.40 |
ENSMUST00000102481.4
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr2_-_34262012 | 1.38 |
ENSMUST00000113132.9
ENSMUST00000040638.15 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr1_+_164624200 | 1.38 |
ENSMUST00000027861.6
|
Dpt
|
dermatopontin |
| chr4_-_104967032 | 1.37 |
ENSMUST00000030243.8
|
Prkaa2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr11_-_61610772 | 1.37 |
ENSMUST00000151780.8
ENSMUST00000148584.2 |
Slc5a10
|
solute carrier family 5 (sodium/glucose cotransporter), member 10 |
| chr14_+_28226697 | 1.37 |
ENSMUST00000063465.12
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr9_-_54467419 | 1.37 |
ENSMUST00000041901.7
|
Cib2
|
calcium and integrin binding family member 2 |
| chr1_-_192955407 | 1.36 |
ENSMUST00000009777.4
|
G0s2
|
G0/G1 switch gene 2 |
| chrX_+_156485570 | 1.36 |
ENSMUST00000112520.2
|
Smpx
|
small muscle protein, X-linked |
| chr16_+_34815177 | 1.35 |
ENSMUST00000231589.2
|
Mylk
|
myosin, light polypeptide kinase |
| chr11_+_116547932 | 1.34 |
ENSMUST00000116318.3
|
Prcd
|
photoreceptor disc component |
| chr11_+_108271990 | 1.34 |
ENSMUST00000146050.2
ENSMUST00000152958.8 |
Apoh
|
apolipoprotein H |
| chrX_+_55824797 | 1.33 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr8_-_105350881 | 1.33 |
ENSMUST00000211903.2
|
Cdh16
|
cadherin 16 |
| chr19_-_32173824 | 1.32 |
ENSMUST00000151822.2
|
Sgms1
|
sphingomyelin synthase 1 |
| chr4_+_95467701 | 1.32 |
ENSMUST00000150830.2
ENSMUST00000134012.9 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr6_+_30509826 | 1.31 |
ENSMUST00000031797.11
|
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr15_-_82291372 | 1.30 |
ENSMUST00000230198.2
ENSMUST00000230248.2 ENSMUST00000072776.5 ENSMUST00000229911.2 |
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr2_+_103254465 | 1.29 |
ENSMUST00000171693.8
|
Elf5
|
E74-like factor 5 |
| chr11_+_76795346 | 1.28 |
ENSMUST00000072633.4
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr2_-_154734824 | 1.28 |
ENSMUST00000099173.11
|
Eif2s2
|
eukaryotic translation initiation factor 2, subunit 2 (beta) |
| chr12_+_85793313 | 1.27 |
ENSMUST00000040461.4
|
Flvcr2
|
feline leukemia virus subgroup C cellular receptor 2 |
| chr2_+_103254401 | 1.26 |
ENSMUST00000028609.14
|
Elf5
|
E74-like factor 5 |
| chr3_+_151143524 | 1.26 |
ENSMUST00000046977.12
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr11_+_76795292 | 1.24 |
ENSMUST00000142166.8
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr13_-_92931317 | 1.23 |
ENSMUST00000022213.8
|
Thbs4
|
thrombospondin 4 |
| chr7_-_5128936 | 1.22 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr9_-_15212849 | 1.22 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr18_-_68562385 | 1.21 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr11_+_67061837 | 1.21 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr19_-_46661321 | 1.20 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr10_-_59277570 | 1.18 |
ENSMUST00000009798.5
|
Oit3
|
oncoprotein induced transcript 3 |
| chr18_-_56695288 | 1.17 |
ENSMUST00000170309.8
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr1_-_74040723 | 1.17 |
ENSMUST00000190389.7
|
Tns1
|
tensin 1 |
| chr4_-_76262464 | 1.16 |
ENSMUST00000050757.16
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr18_-_56695259 | 1.16 |
ENSMUST00000171844.3
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr4_-_82423944 | 1.16 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr14_-_51134930 | 1.16 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr1_-_55127312 | 1.16 |
ENSMUST00000127861.8
ENSMUST00000144077.3 |
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr8_+_93084253 | 1.14 |
ENSMUST00000210246.2
ENSMUST00000034184.12 |
Irx5
|
Iroquois homeobox 5 |
| chr2_-_110144869 | 1.13 |
ENSMUST00000133608.2
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr6_+_82018604 | 1.12 |
ENSMUST00000042974.15
|
Eva1a
|
eva-1 homolog A (C. elegans) |
| chr2_+_90613574 | 1.11 |
ENSMUST00000037206.11
|
Agbl2
|
ATP/GTP binding protein-like 2 |
| chr5_+_8710059 | 1.09 |
ENSMUST00000047753.5
|
Abcb1a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1A |
| chr5_-_35683035 | 1.09 |
ENSMUST00000038676.7
|
Cpz
|
carboxypeptidase Z |
| chr2_+_127205117 | 1.09 |
ENSMUST00000104934.2
|
Adra2b
|
adrenergic receptor, alpha 2b |
| chr9_-_110886306 | 1.08 |
ENSMUST00000195968.2
ENSMUST00000111888.3 |
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr1_-_172722589 | 1.08 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr3_+_96069271 | 1.08 |
ENSMUST00000054356.16
|
Mtmr11
|
myotubularin related protein 11 |
| chr7_+_92210348 | 1.08 |
ENSMUST00000032842.13
ENSMUST00000085017.5 |
Ccdc90b
|
coiled-coil domain containing 90B |
| chr16_+_44913974 | 1.06 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr11_+_72326337 | 1.06 |
ENSMUST00000076443.10
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr2_+_91086299 | 1.06 |
ENSMUST00000134699.8
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr19_-_28945194 | 1.06 |
ENSMUST00000162110.8
|
Spata6l
|
spermatogenesis associated 6 like |
| chr2_-_38604503 | 1.05 |
ENSMUST00000028084.5
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr18_-_35855383 | 1.05 |
ENSMUST00000133064.8
|
Ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr5_+_89034666 | 1.05 |
ENSMUST00000148750.8
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chr3_+_57332735 | 1.05 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr1_-_51955054 | 1.04 |
ENSMUST00000018561.14
ENSMUST00000114537.9 |
Myo1b
|
myosin IB |
| chr6_+_18170686 | 1.04 |
ENSMUST00000045706.12
|
Cftr
|
cystic fibrosis transmembrane conductance regulator |
| chr1_+_171265103 | 1.04 |
ENSMUST00000043839.5
|
F11r
|
F11 receptor |
| chr17_-_35178208 | 1.04 |
ENSMUST00000172753.2
|
Hspa1b
|
heat shock protein 1B |
| chr18_-_38345010 | 1.04 |
ENSMUST00000159405.3
ENSMUST00000160721.8 |
Pcdh1
|
protocadherin 1 |
| chr19_-_10581622 | 1.03 |
ENSMUST00000037678.7
|
Tkfc
|
triokinase, FMN cyclase |
| chr4_+_43414696 | 1.03 |
ENSMUST00000131668.3
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr1_-_51955126 | 1.03 |
ENSMUST00000046390.14
|
Myo1b
|
myosin IB |
| chr13_-_56696222 | 1.02 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr2_+_152468850 | 1.02 |
ENSMUST00000000369.4
ENSMUST00000150913.2 |
Rem1
|
rad and gem related GTP binding protein 1 |
| chr17_-_46749320 | 1.01 |
ENSMUST00000233575.2
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr7_-_126408280 | 1.01 |
ENSMUST00000207534.3
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_-_79382573 | 1.00 |
ENSMUST00000205747.2
|
Plin1
|
perilipin 1 |
| chr9_+_71123061 | 1.00 |
ENSMUST00000034723.6
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chr5_+_96941312 | 0.99 |
ENSMUST00000198631.2
|
Anxa3
|
annexin A3 |
| chr3_+_63203235 | 0.99 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr5_-_87074380 | 0.99 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0061723 | glycophagy(GO:0061723) |
| 0.9 | 7.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.9 | 7.9 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.8 | 2.5 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.8 | 2.5 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.8 | 3.9 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.8 | 2.4 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) positive regulation of establishment of Sertoli cell barrier(GO:1904446) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.8 | 3.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.7 | 2.2 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.6 | 2.5 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.6 | 1.7 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.6 | 4.6 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.6 | 1.7 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.6 | 1.7 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.5 | 4.6 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 1.5 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.5 | 3.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.5 | 1.4 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.5 | 2.7 | GO:0032430 | diacylglycerol biosynthetic process(GO:0006651) positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.5 | 1.4 | GO:0060599 | chemorepulsion of dopaminergic neuron axon(GO:0036518) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) melanocyte proliferation(GO:0097325) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.4 | 1.7 | GO:0046340 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) diacylglycerol catabolic process(GO:0046340) |
| 0.4 | 1.2 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.4 | 1.6 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.4 | 3.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 2.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.4 | 2.3 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.4 | 5.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.4 | 4.1 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.4 | 1.8 | GO:0046618 | drug export(GO:0046618) |
| 0.4 | 1.8 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.4 | 2.6 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.4 | 1.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.4 | 1.8 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.4 | 1.8 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.3 | 1.7 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 1.0 | GO:0097037 | heme export(GO:0097037) |
| 0.3 | 2.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.3 | 1.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.3 | 1.9 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.3 | 0.6 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.3 | 0.9 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.3 | 0.9 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.3 | 3.0 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 0.6 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.3 | 6.2 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.3 | 1.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 0.8 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.3 | 1.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 1.6 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.3 | 6.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 3.3 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.2 | 0.9 | GO:0003360 | brainstem development(GO:0003360) |
| 0.2 | 0.9 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.2 | 4.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 0.4 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.2 | 0.7 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.2 | 1.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.2 | 1.0 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.2 | 1.0 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 2.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 2.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 4.0 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.2 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 1.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.2 | 2.8 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.2 | 2.7 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.2 | 1.1 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 | 6.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 0.5 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.2 | 1.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.2 | 4.7 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.2 | 1.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.8 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 0.7 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.2 | 0.5 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 2.2 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.2 | 1.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.2 | 2.8 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.2 | 1.9 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 0.8 | GO:0070295 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.2 | 1.0 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.2 | 0.5 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.2 | 1.5 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 0.9 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 0.5 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.1 | 1.6 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 2.3 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 0.1 | GO:0021679 | cerebellar molecular layer development(GO:0021679) |
| 0.1 | 1.7 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.1 | 1.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 1.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.9 | GO:0052805 | imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 0.8 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.7 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.1 | 1.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 1.4 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.1 | 0.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.5 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 2.2 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
| 0.1 | 0.7 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 1.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 2.3 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.1 | 0.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.8 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 0.8 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) sulfur oxidation(GO:0019417) |
| 0.1 | 0.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 2.4 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 0.9 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 1.8 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.8 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.5 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.4 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.1 | 0.8 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.4 | GO:0043465 | regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.1 | 3.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.3 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 1.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.3 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 | 0.5 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.4 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 3.6 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.1 | 4.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.3 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.1 | 0.2 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 1.0 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.8 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.5 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 1.0 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 4.3 | GO:1902750 | negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.1 | 3.8 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.1 | 1.8 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.1 | 0.3 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 0.5 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.8 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 0.7 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.8 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 1.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.2 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) lysosomal membrane organization(GO:0097212) positive regulation of protein folding(GO:1903334) |
| 0.1 | 0.5 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.4 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.1 | 0.5 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 2.6 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 0.7 | GO:1903753 | positive regulation of NK T cell activation(GO:0051135) negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 | 1.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 3.1 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.1 | 2.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 1.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.6 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 1.0 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.7 | GO:1904152 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) regulation of retrograde protein transport, ER to cytosol(GO:1904152) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.9 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 0.9 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 1.8 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.2 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.1 | 0.5 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.1 | 0.9 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.1 | 0.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.2 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.1 | 0.6 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.2 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.2 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 1.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 1.2 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.8 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.9 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 1.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 2.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.3 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 1.9 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.5 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 1.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.3 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.3 | GO:0043578 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 2.3 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.6 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.7 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:1903960 | negative regulation of anion channel activity(GO:0010360) negative regulation of anion transmembrane transport(GO:1903960) |
| 0.0 | 0.8 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 9.7 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.5 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 1.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.3 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.6 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 1.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.8 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.8 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 1.2 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 3.5 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.3 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 1.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.2 | GO:0072513 | otic vesicle morphogenesis(GO:0071600) positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.3 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.9 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 4.6 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.0 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 1.8 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.9 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.2 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.4 | GO:0031280 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.2 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.5 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.3 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.0 | 0.3 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.7 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.3 | GO:0035137 | hindlimb morphogenesis(GO:0035137) |
| 0.0 | 0.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.3 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 1.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.9 | 2.7 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.8 | 4.6 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.6 | 2.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.4 | 7.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 2.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.3 | 2.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 1.0 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.3 | 2.4 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.3 | 1.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.3 | 2.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 2.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 1.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 4.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 0.9 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.2 | 0.9 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 1.6 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.7 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 2.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.7 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 3.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 2.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 2.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 2.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.6 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.5 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 0.3 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.1 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.9 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.4 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 7.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 7.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 1.7 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 1.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 8.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0060473 | cortical granule(GO:0060473) |
| 0.0 | 5.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.0 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 2.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.8 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 2.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 3.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.4 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 1.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 3.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 2.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 2.3 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.6 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 5.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.6 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 4.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.6 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 1.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.0 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.0 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 2.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.9 | 2.8 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.8 | 4.2 | GO:2001069 | glycogen binding(GO:2001069) starch binding(GO:2001070) |
| 0.8 | 2.4 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.8 | 4.6 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.7 | 3.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.7 | 3.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.7 | 2.0 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.6 | 2.6 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.6 | 2.5 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.6 | 2.4 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.6 | 2.3 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.5 | 1.5 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.5 | 2.5 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.5 | 6.2 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.5 | 4.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.5 | 2.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.5 | 1.4 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.4 | 3.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.4 | 1.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.4 | 1.6 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.4 | 3.2 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.4 | 2.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.4 | 7.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 1.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.4 | 2.5 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.3 | 1.0 | GO:0004371 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.3 | 1.7 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.3 | 2.0 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 5.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 1.9 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.3 | 2.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.3 | 2.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.3 | 4.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 2.8 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 3.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.3 | 1.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.3 | 1.6 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.3 | 0.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 1.8 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 1.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 1.5 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 1.7 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.2 | 9.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 0.9 | GO:0047726 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.2 | 0.9 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 0.7 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.8 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 1.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 0.6 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.2 | 2.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 1.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 1.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 0.7 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.2 | 3.8 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 5.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 1.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 2.8 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 0.5 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.2 | 1.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.2 | 4.0 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 0.8 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 0.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.2 | 1.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 2.7 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 6.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 1.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 2.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 0.8 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.2 | 0.9 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.2 | 1.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 8.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.9 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 2.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 4.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 2.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.9 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.3 | GO:0004512 | inositol-3-phosphate synthase activity(GO:0004512) intramolecular lyase activity(GO:0016872) |
| 0.1 | 0.8 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 1.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.7 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 1.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 3.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 12.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 1.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.5 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.7 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.6 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 2.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.0 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 1.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 2.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.3 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 2.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 2.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 3.1 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.9 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 1.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.8 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 2.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 2.5 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 3.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.2 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 1.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 1.6 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.2 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 2.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.7 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 1.1 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 7.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 6.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 3.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 5.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 5.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 7.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 6.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 8.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 3.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.5 | 6.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 3.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 10.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 2.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 2.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 9.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 3.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 6.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 9.9 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 4.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 3.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 3.7 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.1 | 2.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.8 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 0.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 3.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 6.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 6.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 2.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 2.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.1 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 2.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 3.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 2.4 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.3 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |