Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
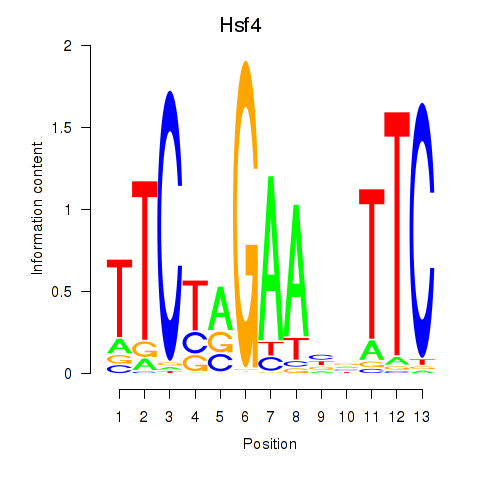
Results for Hsf4
Z-value: 1.52
Transcription factors associated with Hsf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf4
|
ENSMUSG00000033249.11 | Hsf4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf4 | mm39_v1_chr8_+_105996469_105996506 | -0.57 | 1.4e-07 | Click! |
Activity profile of Hsf4 motif
Sorted Z-values of Hsf4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_28544356 | 10.66 |
ENSMUST00000060409.13
ENSMUST00000056097.11 ENSMUST00000105516.9 |
Themis
|
thymocyte selection associated |
| chr2_+_172821620 | 10.56 |
ENSMUST00000109125.8
ENSMUST00000109126.5 ENSMUST00000050442.15 |
Spo11
|
SPO11 initiator of meiotic double stranded breaks |
| chr4_-_107541421 | 9.91 |
ENSMUST00000069271.5
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr7_+_43625387 | 9.68 |
ENSMUST00000081399.4
|
Klk1b9
|
kallikrein 1-related peptidase b9 |
| chr11_+_46295547 | 9.22 |
ENSMUST00000063166.6
|
Fam71b
|
family with sequence similarity 71, member B |
| chr5_+_136721938 | 8.85 |
ENSMUST00000196068.5
ENSMUST00000005611.10 |
Myl10
|
myosin, light chain 10, regulatory |
| chr1_-_166066298 | 8.73 |
ENSMUST00000038782.4
ENSMUST00000194057.6 |
Mael
|
maelstrom spermatogenic transposon silencer |
| chr15_-_76259126 | 8.58 |
ENSMUST00000071119.8
|
Tssk5
|
testis-specific serine kinase 5 |
| chr15_-_74671382 | 8.51 |
ENSMUST00000168815.8
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr1_+_165312738 | 8.49 |
ENSMUST00000111440.8
ENSMUST00000027852.15 ENSMUST00000111439.8 |
Adcy10
|
adenylate cyclase 10 |
| chr17_+_34636321 | 8.19 |
ENSMUST00000142317.8
|
BC051142
|
cDNA sequence BC051142 |
| chr2_-_152422220 | 7.97 |
ENSMUST00000053180.4
|
Defb19
|
defensin beta 19 |
| chr15_-_74671739 | 7.86 |
ENSMUST00000060301.6
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr11_+_104576528 | 7.71 |
ENSMUST00000148007.3
ENSMUST00000212287.2 |
Gm11639
|
predicted gene 11639 |
| chr2_+_144665576 | 7.70 |
ENSMUST00000028918.4
|
Scp2d1
|
SCP2 sterol-binding domain containing 1 |
| chr4_-_46413484 | 7.64 |
ENSMUST00000071096.3
|
Hemgn
|
hemogen |
| chr14_+_54457978 | 7.52 |
ENSMUST00000103740.2
ENSMUST00000198398.5 |
Trac
|
T cell receptor alpha constant |
| chr14_+_54380308 | 7.51 |
ENSMUST00000196323.2
|
Trdc
|
T cell receptor delta, constant region |
| chr2_+_172314433 | 7.34 |
ENSMUST00000029007.3
|
Fam209
|
family with sequence similarity 209 |
| chr11_-_101490011 | 7.03 |
ENSMUST00000209862.2
ENSMUST00000154147.2 |
Gm11634
|
predicted gene 11634 |
| chr11_+_82802079 | 7.03 |
ENSMUST00000018989.14
ENSMUST00000164945.3 |
Unc45b
|
unc-45 myosin chaperone B |
| chr10_+_100428212 | 7.01 |
ENSMUST00000187119.7
ENSMUST00000188736.7 ENSMUST00000191336.7 |
1700017N19Rik
|
RIKEN cDNA 1700017N19 gene |
| chr15_+_81943345 | 6.98 |
ENSMUST00000100396.4
|
4930407I10Rik
|
RIKEN cDNA 4930407I10 gene |
| chr2_+_121991181 | 6.97 |
ENSMUST00000036089.8
|
Trim69
|
tripartite motif-containing 69 |
| chr17_+_28749808 | 6.96 |
ENSMUST00000233837.2
ENSMUST00000025060.4 |
Armc12
|
armadillo repeat containing 12 |
| chr17_+_48607405 | 6.94 |
ENSMUST00000170941.3
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr2_+_103953083 | 6.85 |
ENSMUST00000040374.6
|
A930018P22Rik
|
RIKEN cDNA A930018P22 gene |
| chr10_+_100428246 | 6.64 |
ENSMUST00000041162.13
ENSMUST00000190386.7 ENSMUST00000190708.7 |
1700017N19Rik
|
RIKEN cDNA 1700017N19 gene |
| chr12_+_71158192 | 6.51 |
ENSMUST00000021482.6
|
Tomm20l
|
translocase of outer mitochondrial membrane 20-like |
| chr17_+_28749780 | 6.45 |
ENSMUST00000233923.2
|
Armc12
|
armadillo repeat containing 12 |
| chr17_+_27248233 | 6.42 |
ENSMUST00000053683.7
ENSMUST00000236222.2 |
Ggnbp1
|
gametogenetin binding protein 1 |
| chr11_+_101877876 | 6.37 |
ENSMUST00000010985.8
|
Cfap97d1
|
CFAP97 domain containing 1 |
| chr17_+_31783708 | 6.36 |
ENSMUST00000097352.11
ENSMUST00000237248.2 ENSMUST00000235869.2 ENSMUST00000175806.9 |
Pknox1
|
Pbx/knotted 1 homeobox |
| chrY_+_17400761 | 6.32 |
ENSMUST00000179408.3
|
Rbm31y
|
RNA binding motif 31, Y-linked |
| chr1_-_189420270 | 6.20 |
ENSMUST00000171929.8
ENSMUST00000165962.2 |
Cenpf
|
centromere protein F |
| chr17_-_34218301 | 6.09 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr18_-_9619460 | 6.06 |
ENSMUST00000234003.2
ENSMUST00000062769.7 |
Cetn1
|
centrin 1 |
| chr2_+_148631276 | 6.05 |
ENSMUST00000168443.8
ENSMUST00000028932.4 |
Cst12
|
cystatin 12 |
| chr17_+_87224776 | 6.02 |
ENSMUST00000042172.7
|
Tmem247
|
transmembrane protein 247 |
| chr5_-_92822984 | 5.98 |
ENSMUST00000060930.10
|
Ccdc158
|
coiled-coil domain containing 158 |
| chr19_+_8718073 | 5.97 |
ENSMUST00000163172.2
|
1700092M07Rik
|
RIKEN cDNA 1700092M07 gene |
| chr9_+_106377181 | 5.95 |
ENSMUST00000085114.8
|
Iqcf1
|
IQ motif containing F1 |
| chr17_-_57181420 | 5.89 |
ENSMUST00000043062.5
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr5_+_107977965 | 5.89 |
ENSMUST00000072578.8
|
Ube2d2b
|
ubiquitin-conjugating enzyme E2D 2B |
| chr9_+_106377153 | 5.85 |
ENSMUST00000164965.3
|
Iqcf1
|
IQ motif containing F1 |
| chr2_-_172212426 | 5.82 |
ENSMUST00000109139.8
ENSMUST00000028997.8 ENSMUST00000109140.10 |
Aurka
|
aurora kinase A |
| chr1_-_156168522 | 5.76 |
ENSMUST00000212747.2
|
Axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr7_+_43701714 | 5.76 |
ENSMUST00000079859.7
|
Klk1b27
|
kallikrein 1-related peptidase b27 |
| chr7_-_119122681 | 5.67 |
ENSMUST00000033267.4
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr8_-_11528615 | 5.40 |
ENSMUST00000033900.7
|
Rab20
|
RAB20, member RAS oncogene family |
| chr7_-_133203838 | 5.36 |
ENSMUST00000033275.4
|
Tex36
|
testis expressed 36 |
| chr5_-_107873883 | 5.33 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr17_+_47904355 | 5.31 |
ENSMUST00000182209.8
|
Ccnd3
|
cyclin D3 |
| chrX_+_56454507 | 5.29 |
ENSMUST00000114725.3
|
Tm9sf5
|
transmembrane 9 superfamily member 5 |
| chr17_+_35643818 | 5.27 |
ENSMUST00000174699.8
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr14_-_66071412 | 5.23 |
ENSMUST00000022613.10
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr7_+_43600038 | 5.22 |
ENSMUST00000072204.5
|
Klk1b8
|
kallikrein 1-related peptidase b8 |
| chr18_+_70605722 | 5.22 |
ENSMUST00000174118.8
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr17_+_48606948 | 5.20 |
ENSMUST00000233092.2
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr10_+_130158737 | 5.17 |
ENSMUST00000217702.2
ENSMUST00000042586.10 ENSMUST00000218605.2 |
Tespa1
|
thymocyte expressed, positive selection associated 1 |
| chr1_-_128520002 | 5.17 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr18_+_70605691 | 5.14 |
ENSMUST00000164223.8
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr19_+_25482982 | 5.13 |
ENSMUST00000025755.11
|
Dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr4_-_41048124 | 5.07 |
ENSMUST00000030136.13
|
Aqp7
|
aquaporin 7 |
| chr17_+_35658131 | 5.07 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr17_+_35780977 | 5.06 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chrX_+_160080789 | 5.02 |
ENSMUST00000207853.2
ENSMUST00000207800.2 |
Gm15262
|
predicted gene 15262 |
| chr3_-_146357059 | 4.99 |
ENSMUST00000149825.2
ENSMUST00000049703.6 |
4930503B20Rik
|
RIKEN cDNA 4930503B20 gene |
| chr15_+_88484484 | 4.98 |
ENSMUST00000066949.9
|
Zdhhc25
|
zinc finger, DHHC domain containing 25 |
| chr8_-_106706035 | 4.97 |
ENSMUST00000034371.9
|
Dpep3
|
dipeptidase 3 |
| chr1_-_156248442 | 4.96 |
ENSMUST00000179985.8
ENSMUST00000179744.2 |
Axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr11_+_80749184 | 4.95 |
ENSMUST00000103223.8
ENSMUST00000103222.4 |
Spaca3
|
sperm acrosome associated 3 |
| chr11_-_70510010 | 4.94 |
ENSMUST00000102556.10
ENSMUST00000014753.9 |
Chrne
|
cholinergic receptor, nicotinic, epsilon polypeptide |
| chr11_-_118021460 | 4.89 |
ENSMUST00000132685.9
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr5_+_92479677 | 4.85 |
ENSMUST00000154245.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chrX_-_133377225 | 4.72 |
ENSMUST00000009740.9
|
Taf7l
|
TATA-box binding protein associated factor 7 like |
| chr18_+_70605630 | 4.71 |
ENSMUST00000168249.9
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr7_-_30292351 | 4.67 |
ENSMUST00000108151.3
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr10_+_76398106 | 4.67 |
ENSMUST00000009259.5
|
Spatc1l
|
spermatogenesis and centriole associated 1 like |
| chr14_-_101437750 | 4.63 |
ENSMUST00000187304.2
|
Prr30
|
proline rich 30 |
| chr12_-_104964936 | 4.63 |
ENSMUST00000109927.2
ENSMUST00000095439.11 |
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr17_-_45903494 | 4.62 |
ENSMUST00000163492.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr11_+_5519677 | 4.62 |
ENSMUST00000109856.8
ENSMUST00000109855.8 ENSMUST00000118112.9 |
Ankrd36
|
ankyrin repeat domain 36 |
| chr18_+_70605476 | 4.62 |
ENSMUST00000114959.9
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr17_+_57182472 | 4.62 |
ENSMUST00000025048.7
|
Acsbg3
|
acyl-CoA synthetase bubblegum family member 3 |
| chr5_+_25386487 | 4.58 |
ENSMUST00000114965.2
|
Galntl5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr6_-_148847854 | 4.58 |
ENSMUST00000139355.8
ENSMUST00000146457.2 ENSMUST00000054080.15 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr6_-_128415640 | 4.57 |
ENSMUST00000032508.11
|
Fkbp4
|
FK506 binding protein 4 |
| chr1_+_37192920 | 4.54 |
ENSMUST00000169057.3
|
Vwa3b
|
von Willebrand factor A domain containing 3B |
| chr12_+_104067346 | 4.52 |
ENSMUST00000021495.4
|
Serpina5
|
serine (or cysteine) peptidase inhibitor, clade A, member 5 |
| chr6_-_129252396 | 4.47 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr19_-_28945194 | 4.45 |
ENSMUST00000162110.8
|
Spata6l
|
spermatogenesis associated 6 like |
| chr11_+_121036969 | 4.44 |
ENSMUST00000039088.9
ENSMUST00000155694.2 |
Tex19.1
|
testis expressed gene 19.1 |
| chr11_+_96355441 | 4.38 |
ENSMUST00000071510.14
ENSMUST00000107662.9 |
Skap1
|
src family associated phosphoprotein 1 |
| chr6_-_133830613 | 4.35 |
ENSMUST00000203168.2
ENSMUST00000048032.5 |
Kap
|
kidney androgen regulated protein |
| chr7_+_43751749 | 4.30 |
ENSMUST00000085455.6
|
Klk1b21
|
kallikrein 1-related peptidase b21 |
| chr5_+_92479687 | 4.30 |
ENSMUST00000125462.8
ENSMUST00000113083.9 ENSMUST00000121096.8 |
Art3
|
ADP-ribosyltransferase 3 |
| chr11_-_116734275 | 4.26 |
ENSMUST00000047616.10
|
Jmjd6
|
jumonji domain containing 6 |
| chr19_+_25483070 | 4.26 |
ENSMUST00000087525.5
|
Dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr2_-_118087401 | 4.15 |
ENSMUST00000231133.2
ENSMUST00000028820.7 ENSMUST00000028821.10 |
Fsip1
|
fibrous sheath-interacting protein 1 |
| chr5_-_92823111 | 4.15 |
ENSMUST00000151180.8
ENSMUST00000150359.2 |
Ccdc158
|
coiled-coil domain containing 158 |
| chr6_+_125026915 | 4.12 |
ENSMUST00000088294.12
ENSMUST00000032481.8 |
Acrbp
|
proacrosin binding protein |
| chr2_-_118558852 | 4.11 |
ENSMUST00000102524.8
|
Plcb2
|
phospholipase C, beta 2 |
| chr13_-_35100960 | 4.08 |
ENSMUST00000225242.2
|
Fam217a
|
family with sequence similarity 217, member A |
| chr8_+_55053809 | 4.06 |
ENSMUST00000033917.7
|
Spata4
|
spermatogenesis associated 4 |
| chr11_+_100510043 | 4.04 |
ENSMUST00000107376.8
|
Nkiras2
|
NFKB inhibitor interacting Ras-like protein 2 |
| chr2_-_118558825 | 4.00 |
ENSMUST00000159756.2
|
Plcb2
|
phospholipase C, beta 2 |
| chr7_+_43847615 | 4.00 |
ENSMUST00000085450.4
|
Klk1b3
|
kallikrein 1-related peptidase b3 |
| chr16_+_4653271 | 3.98 |
ENSMUST00000100211.11
|
4930562C15Rik
|
RIKEN cDNA 4930562C15 gene |
| chr6_-_136918885 | 3.97 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr16_+_8498618 | 3.96 |
ENSMUST00000201722.2
|
Gm5767
|
predicted gene 5767 |
| chr14_+_43116106 | 3.95 |
ENSMUST00000169587.2
|
Gm8126
|
predicted gene 8126 |
| chr11_+_96355413 | 3.93 |
ENSMUST00000103154.11
ENSMUST00000100521.10 ENSMUST00000100519.11 |
Skap1
|
src family associated phosphoprotein 1 |
| chr7_-_46365108 | 3.91 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr9_+_123596276 | 3.90 |
ENSMUST00000166236.9
ENSMUST00000111454.4 ENSMUST00000168910.2 |
Ccr9
|
chemokine (C-C motif) receptor 9 |
| chr11_-_106811507 | 3.90 |
ENSMUST00000103067.10
|
Smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr2_+_119727689 | 3.89 |
ENSMUST00000046717.13
ENSMUST00000079934.12 ENSMUST00000110774.8 ENSMUST00000110773.9 ENSMUST00000156510.2 |
Mga
|
MAX gene associated |
| chr6_-_136918495 | 3.88 |
ENSMUST00000111892.8
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr11_-_89893707 | 3.86 |
ENSMUST00000020864.9
|
Pctp
|
phosphatidylcholine transfer protein |
| chr7_+_43837629 | 3.84 |
ENSMUST00000073713.8
|
Klk1b24
|
kallikrein 1-related peptidase b24 |
| chr6_+_18866308 | 3.84 |
ENSMUST00000031489.10
|
Ankrd7
|
ankyrin repeat domain 7 |
| chr14_-_44408828 | 3.77 |
ENSMUST00000100688.3
|
4930503E14Rik
|
RIKEN cDNA 4930503E14 gene |
| chr17_+_35643853 | 3.74 |
ENSMUST00000113879.4
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr3_+_89269282 | 3.73 |
ENSMUST00000208216.2
|
Dcst2
|
DC-STAMP domain containing 2 |
| chr16_-_55755160 | 3.72 |
ENSMUST00000122280.8
ENSMUST00000121703.3 |
Cep97
|
centrosomal protein 97 |
| chr19_+_56814700 | 3.70 |
ENSMUST00000078723.11
ENSMUST00000121249.8 |
Tdrd1
|
tudor domain containing 1 |
| chr14_+_54455293 | 3.68 |
ENSMUST00000103738.2
|
Traj2
|
T cell receptor alpha joining 2 |
| chr10_+_76398138 | 3.67 |
ENSMUST00000105414.2
|
Spatc1l
|
spermatogenesis and centriole associated 1 like |
| chr10_-_39039790 | 3.65 |
ENSMUST00000076713.6
|
Ccn6
|
cellular communication network factor 6 |
| chr17_+_25381414 | 3.64 |
ENSMUST00000073277.12
ENSMUST00000182621.8 |
Ccdc154
|
coiled-coil domain containing 154 |
| chr6_+_18866339 | 3.63 |
ENSMUST00000115396.7
|
Ankrd7
|
ankyrin repeat domain 7 |
| chr19_+_10611574 | 3.54 |
ENSMUST00000055115.9
|
Vwce
|
von Willebrand factor C and EGF domains |
| chr19_+_4036562 | 3.52 |
ENSMUST00000236224.2
ENSMUST00000236510.2 ENSMUST00000237910.2 ENSMUST00000235612.2 ENSMUST00000054030.8 |
Acy3
|
aspartoacylase (aminoacylase) 3 |
| chr5_+_25386452 | 3.52 |
ENSMUST00000030778.9
|
Galntl5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr1_-_161807205 | 3.50 |
ENSMUST00000162676.2
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chr17_+_47904441 | 3.50 |
ENSMUST00000182874.3
|
Ccnd3
|
cyclin D3 |
| chr6_-_136918671 | 3.49 |
ENSMUST00000032344.12
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr4_-_49408040 | 3.46 |
ENSMUST00000081541.9
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr16_-_22084700 | 3.44 |
ENSMUST00000161286.8
|
Tra2b
|
transformer 2 beta |
| chr6_-_124865155 | 3.39 |
ENSMUST00000024044.7
|
Cd4
|
CD4 antigen |
| chr11_+_70350252 | 3.38 |
ENSMUST00000108563.9
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr12_-_69274936 | 3.35 |
ENSMUST00000221411.2
ENSMUST00000021359.7 |
Pole2
|
polymerase (DNA directed), epsilon 2 (p59 subunit) |
| chr2_+_122478882 | 3.29 |
ENSMUST00000142767.8
|
AA467197
|
expressed sequence AA467197 |
| chr9_-_75317233 | 3.27 |
ENSMUST00000049355.11
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr8_-_22675773 | 3.26 |
ENSMUST00000046916.9
|
Ckap2
|
cytoskeleton associated protein 2 |
| chr9_-_37166699 | 3.22 |
ENSMUST00000161114.2
|
Slc37a2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2 |
| chr7_-_19005721 | 3.19 |
ENSMUST00000032561.9
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr9_+_72345801 | 3.19 |
ENSMUST00000184604.8
ENSMUST00000034746.10 |
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr7_+_49408847 | 3.18 |
ENSMUST00000085272.7
ENSMUST00000207895.2 |
Htatip2
|
HIV-1 Tat interactive protein 2 |
| chr5_+_149121458 | 3.16 |
ENSMUST00000122160.8
ENSMUST00000100410.10 ENSMUST00000119685.8 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chrX_+_20934645 | 3.15 |
ENSMUST00000040437.6
|
Spaca5
|
sperm acrosome associated 5 |
| chr18_+_77152890 | 3.12 |
ENSMUST00000114777.10
|
Pias2
|
protein inhibitor of activated STAT 2 |
| chr19_+_56814634 | 3.12 |
ENSMUST00000111606.8
|
Tdrd1
|
tudor domain containing 1 |
| chr14_+_41295168 | 3.09 |
ENSMUST00000163912.3
ENSMUST00000111874.3 |
1700049E17Rik2
|
tandem duplication of RIKEN cDNA 1700049E17 gene, gene 2 |
| chr5_-_116427003 | 3.09 |
ENSMUST00000086483.4
ENSMUST00000050178.13 |
Ccdc60
|
coiled-coil domain containing 60 |
| chr11_-_69768875 | 3.06 |
ENSMUST00000178597.3
|
Tmem95
|
transmembrane protein 95 |
| chr7_-_114162125 | 3.05 |
ENSMUST00000211506.2
ENSMUST00000119712.8 ENSMUST00000032908.15 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr15_-_59888446 | 3.04 |
ENSMUST00000096421.4
|
4933412E24Rik
|
RIKEN cDNA 4933412E24 gene |
| chr5_+_149121355 | 3.03 |
ENSMUST00000050472.16
|
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr6_+_125026865 | 3.03 |
ENSMUST00000112413.8
|
Acrbp
|
proacrosin binding protein |
| chr14_+_43781033 | 2.98 |
ENSMUST00000169023.2
|
Gm5799
|
predicted gene 5799 |
| chr3_+_108987779 | 2.95 |
ENSMUST00000029478.5
|
Slc25a54
|
solute carrier family 25, member 54 |
| chr17_-_56428968 | 2.94 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr7_-_25239229 | 2.93 |
ENSMUST00000044547.10
ENSMUST00000066503.14 ENSMUST00000064862.13 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr11_+_70350436 | 2.93 |
ENSMUST00000039093.10
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr9_-_13738204 | 2.93 |
ENSMUST00000147115.8
|
Cep57
|
centrosomal protein 57 |
| chr7_+_43874752 | 2.91 |
ENSMUST00000075162.5
|
Klk1
|
kallikrein 1 |
| chr17_+_35598583 | 2.88 |
ENSMUST00000081435.5
|
H2-Q4
|
histocompatibility 2, Q region locus 4 |
| chr14_+_54454749 | 2.86 |
ENSMUST00000103737.3
|
Traj3
|
T cell receptor alpha joining 3 |
| chr10_-_62343516 | 2.81 |
ENSMUST00000020271.13
|
Srgn
|
serglycin |
| chrX_-_159689993 | 2.79 |
ENSMUST00000069417.6
|
Gja6
|
gap junction protein, alpha 6 |
| chrX_-_139714182 | 2.76 |
ENSMUST00000044179.8
|
Tex13b
|
testis expressed 13B |
| chr2_+_130247159 | 2.76 |
ENSMUST00000188900.2
ENSMUST00000110281.2 ENSMUST00000189961.2 |
Gm28372
1700020A23Rik
|
predicted gene 28372 RIKEN cDNA 1700020A23 gene |
| chr7_-_42097503 | 2.74 |
ENSMUST00000032648.5
|
4933421I07Rik
|
RIKEN cDNA 4933421I07 gene |
| chr7_+_138792890 | 2.74 |
ENSMUST00000016124.15
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr11_+_70350725 | 2.71 |
ENSMUST00000147289.2
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr11_+_101473062 | 2.67 |
ENSMUST00000039581.14
ENSMUST00000100403.9 ENSMUST00000107194.8 ENSMUST00000128614.2 |
Tmem106a
|
transmembrane protein 106A |
| chr14_+_43985323 | 2.66 |
ENSMUST00000100643.10
ENSMUST00000179860.2 |
Gm6526
|
predicted gene 6526 |
| chr11_+_116734104 | 2.65 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr14_+_54014387 | 2.63 |
ENSMUST00000103670.4
|
Trav3-4
|
T cell receptor alpha variable 3-4 |
| chr10_-_86843878 | 2.62 |
ENSMUST00000035288.17
|
Stab2
|
stabilin 2 |
| chr4_-_86588267 | 2.62 |
ENSMUST00000000466.13
|
Plin2
|
perilipin 2 |
| chr5_-_87572060 | 2.61 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr5_+_149121488 | 2.60 |
ENSMUST00000139474.8
ENSMUST00000117878.8 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr10_+_4296806 | 2.60 |
ENSMUST00000216139.3
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr18_+_20443795 | 2.59 |
ENSMUST00000077146.4
|
Dsg1a
|
desmoglein 1 alpha |
| chrX_+_41238410 | 2.58 |
ENSMUST00000127618.8
|
Stag2
|
stromal antigen 2 |
| chr14_+_41746906 | 2.58 |
ENSMUST00000095985.5
|
1700049E17Rik1
|
RIKEN cDNA 1700049E17 gene, gene 1 |
| chr5_-_88823989 | 2.57 |
ENSMUST00000078945.12
|
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chrX_+_41238193 | 2.57 |
ENSMUST00000115073.9
ENSMUST00000115072.8 |
Stag2
|
stromal antigen 2 |
| chr11_-_49077986 | 2.54 |
ENSMUST00000046522.13
|
Btnl9
|
butyrophilin-like 9 |
| chr14_+_41000762 | 2.54 |
ENSMUST00000172412.2
|
Gm2832
|
predicted gene 2832 |
| chr1_+_130793406 | 2.53 |
ENSMUST00000038829.7
|
Fcmr
|
Fc fragment of IgM receptor |
| chr8_-_71990085 | 2.53 |
ENSMUST00000051672.9
|
Bst2
|
bone marrow stromal cell antigen 2 |
| chrY_-_17937330 | 2.51 |
ENSMUST00000179900.2
|
Srsy
|
serine-rich, secreted, Y-linked |
| chr1_+_69866145 | 2.49 |
ENSMUST00000065425.12
ENSMUST00000113940.4 |
Spag16
|
sperm associated antigen 16 |
| chr11_+_96355475 | 2.49 |
ENSMUST00000107663.4
|
Skap1
|
src family associated phosphoprotein 1 |
| chr6_-_136918844 | 2.46 |
ENSMUST00000204934.2
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr6_+_48624295 | 2.44 |
ENSMUST00000078223.6
ENSMUST00000203509.2 |
Gimap8
|
GTPase, IMAP family member 8 |
| chr9_-_36637923 | 2.44 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 2.0 | 11.8 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 1.9 | 9.7 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 1.9 | 5.8 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 1.8 | 8.8 | GO:0030576 | Cajal body organization(GO:0030576) |
| 1.7 | 5.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 1.6 | 4.7 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 1.5 | 13.8 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 1.5 | 7.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.5 | 4.4 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 1.3 | 5.3 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 1.3 | 3.9 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 1.2 | 6.2 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 1.2 | 5.9 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.1 | 11.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 1.1 | 5.5 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 1.1 | 3.2 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 1.0 | 5.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 1.0 | 5.1 | GO:0070295 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.9 | 2.8 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.9 | 6.6 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.8 | 3.3 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.7 | 15.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.7 | 2.8 | GO:0033370 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.7 | 3.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.7 | 4.6 | GO:0015862 | uridine transport(GO:0015862) |
| 0.6 | 1.9 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.6 | 1.9 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.6 | 1.8 | GO:0090265 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.6 | 3.4 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.6 | 3.4 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.5 | 3.2 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.5 | 4.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.5 | 2.6 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.5 | 5.8 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.5 | 1.6 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.5 | 12.0 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.5 | 1.5 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.5 | 5.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.5 | 2.5 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.5 | 3.9 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.5 | 2.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.4 | 8.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.4 | 22.0 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.4 | 3.1 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.4 | 2.5 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.4 | 1.3 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.4 | 8.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.4 | 2.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.4 | 1.1 | GO:0006530 | asparagine catabolic process(GO:0006530) |
| 0.4 | 1.1 | GO:0061762 | protein lipidation involved in autophagosome assembly(GO:0061739) CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.4 | 2.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 3.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 8.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.3 | 6.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.3 | 1.0 | GO:0097212 | protein targeting to vacuole involved in autophagy(GO:0071211) lysosomal membrane organization(GO:0097212) positive regulation of protein folding(GO:1903334) |
| 0.3 | 4.6 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.3 | 1.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.3 | 0.9 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.3 | 2.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.3 | 2.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.3 | 2.9 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 2.0 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.3 | 12.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.3 | 1.4 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.3 | 1.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.3 | 0.8 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.3 | 0.8 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.3 | 10.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 0.9 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 2.3 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.2 | 0.4 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.2 | 1.3 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.2 | 2.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.2 | 41.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 2.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 1.9 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 0.4 | GO:0046618 | drug export(GO:0046618) |
| 0.2 | 1.6 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.2 | 3.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 3.0 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 4.6 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 | 2.8 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 1.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 9.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.2 | 2.0 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.2 | 0.5 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.2 | 1.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 2.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 2.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 1.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 1.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 2.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 1.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.2 | 3.5 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.2 | 3.9 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 0.3 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 | 2.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.9 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.1 | 0.6 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 11.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.4 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.6 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.9 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 2.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 3.1 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.1 | 5.2 | GO:0007140 | male meiosis(GO:0007140) |
| 0.1 | 0.4 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 2.5 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 8.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 1.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 1.5 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.6 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 4.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 5.1 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.1 | 4.9 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 2.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 2.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.4 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 2.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 6.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 1.6 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.7 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 1.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 1.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 1.2 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.1 | 1.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 1.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.7 | GO:0021995 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 0.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 6.0 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 0.7 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.5 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 1.1 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 2.3 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.6 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.4 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 | 2.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 2.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 1.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.4 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 23.7 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 1.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.6 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 4.3 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 7.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.9 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.1 | 2.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 5.4 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 2.2 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 12.2 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 1.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.9 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 1.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 1.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 5.5 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 1.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 3.3 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 3.2 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 1.0 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.0 | 0.2 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.5 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.0 | 1.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.7 | GO:0010524 | positive regulation of calcium ion transport into cytosol(GO:0010524) |
| 0.0 | 0.6 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.2 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.4 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 31.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 2.0 | 8.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.5 | 8.7 | GO:0071547 | piP-body(GO:0071547) |
| 1.1 | 6.6 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 1.1 | 3.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 1.0 | 5.8 | GO:0032133 | chromosome passenger complex(GO:0032133) germinal vesicle(GO:0042585) |
| 0.9 | 2.6 | GO:0060473 | cortical granule(GO:0060473) |
| 0.9 | 6.8 | GO:0071546 | pi-body(GO:0071546) |
| 0.7 | 2.0 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.7 | 3.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.6 | 2.5 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.6 | 2.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.5 | 6.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.5 | 4.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 13.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.4 | 6.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.4 | 4.5 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.4 | 4.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 9.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 2.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.3 | 2.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 1.9 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 3.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 1.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 4.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 2.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 2.9 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 0.6 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.2 | 1.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 10.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 3.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 2.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 1.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 5.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 29.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 6.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 23.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.8 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 17.1 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 1.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 5.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 3.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 5.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 2.7 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 1.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 3.4 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 2.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 3.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 14.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 2.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 6.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 7.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 6.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 2.0 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 2.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 2.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 5.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 3.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.6 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 2.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 3.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 6.5 | GO:0000228 | nuclear chromosome(GO:0000228) |
| 0.0 | 4.0 | GO:0098687 | chromosomal region(GO:0098687) |
| 0.0 | 13.4 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 7.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 11.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 6.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 8.5 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 1.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 3.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0070138 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 2.6 | 10.6 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 2.2 | 6.5 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 1.9 | 31.8 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 1.5 | 13.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.5 | 4.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 1.2 | 4.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.0 | 8.3 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.9 | 4.6 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.9 | 2.6 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.8 | 3.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.7 | 5.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.7 | 4.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.7 | 7.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.7 | 6.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.7 | 6.6 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.7 | 9.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.6 | 1.9 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.6 | 1.8 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.6 | 3.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.6 | 7.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.6 | 1.7 | GO:0005333 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.6 | 2.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.5 | 1.5 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.5 | 3.9 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.5 | 2.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.4 | 8.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.4 | 3.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.4 | 3.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.4 | 2.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 4.7 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.4 | 1.1 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.4 | 0.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.4 | 39.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.3 | 1.4 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 25.7 | GO:0032934 | sterol binding(GO:0032934) |
| 0.3 | 1.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.3 | 8.1 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.3 | 2.3 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.3 | 3.9 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.3 | 2.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.3 | 1.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.3 | 8.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.3 | 1.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 3.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 5.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.3 | 5.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 4.9 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.3 | 2.9 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.3 | 2.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 1.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.2 | 3.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 1.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 7.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 1.6 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 1.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 1.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 1.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.2 | 3.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 3.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 2.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 2.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 2.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 10.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 5.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 8.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 4.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.6 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.0 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 7.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 3.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 5.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 2.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 8.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 3.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 2.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 3.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 4.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 3.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.4 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 1.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 2.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 2.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 3.2 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.1 | 2.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 7.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.9 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 5.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.5 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 3.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 4.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 3.8 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 7.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 2.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 5.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 22.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 4.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.7 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 14.8 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.5 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 2.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.6 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 1.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 2.0 | GO:0008168 | methyltransferase activity(GO:0008168) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 8.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 6.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 6.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 10.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 10.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 6.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 13.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.9 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 4.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 5.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 3.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 2.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 4.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 4.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.6 | 8.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.6 | 25.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.5 | 1.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.5 | 4.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.5 | 2.8 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.4 | 5.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 4.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 13.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.3 | 4.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 6.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 3.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.2 | 6.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 1.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 1.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 4.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 8.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 5.1 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 2.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 7.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 2.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 4.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 4.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 4.2 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 2.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 2.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.8 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 2.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.6 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |