Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
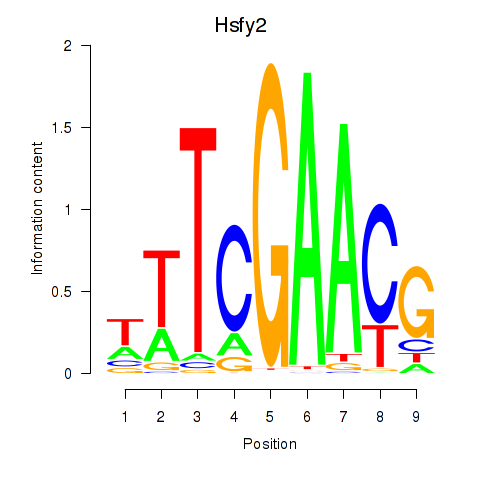
Results for Hsfy2
Z-value: 1.02
Transcription factors associated with Hsfy2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsfy2
|
ENSMUSG00000045336.6 | Hsfy2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsfy2 | mm39_v1_chr1_-_56676589_56676606 | 0.40 | 5.4e-04 | Click! |
Activity profile of Hsfy2 motif
Sorted Z-values of Hsfy2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsfy2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_44893077 | 6.66 |
ENSMUST00000034602.9
|
Cd3d
|
CD3 antigen, delta polypeptide |
| chr9_-_57552392 | 6.25 |
ENSMUST00000216934.2
|
Csk
|
c-src tyrosine kinase |
| chr2_+_152689913 | 5.90 |
ENSMUST00000028969.9
|
Tpx2
|
TPX2, microtubule-associated |
| chr2_+_152689881 | 5.60 |
ENSMUST00000164120.8
ENSMUST00000178997.8 ENSMUST00000109816.8 |
Tpx2
|
TPX2, microtubule-associated |
| chr11_+_11636213 | 5.14 |
ENSMUST00000076700.11
ENSMUST00000048122.13 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr11_-_102815910 | 5.05 |
ENSMUST00000021311.10
|
Kif18b
|
kinesin family member 18B |
| chr6_+_41039255 | 4.59 |
ENSMUST00000103266.3
|
Trbv5
|
T cell receptor beta, variable 5 |
| chr1_+_139382485 | 4.54 |
ENSMUST00000200083.5
ENSMUST00000053364.12 |
Aspm
|
abnormal spindle microtubule assembly |
| chr13_+_51799268 | 4.29 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr8_+_4375212 | 3.83 |
ENSMUST00000127460.8
ENSMUST00000136191.8 |
Ccl25
|
chemokine (C-C motif) ligand 25 |
| chr9_-_62189441 | 3.76 |
ENSMUST00000056949.5
|
Spesp1
|
sperm equatorial segment protein 1 |
| chr7_-_140793990 | 3.75 |
ENSMUST00000026573.7
ENSMUST00000170841.9 |
Lmntd2
|
lamin tail domain containing 2 |
| chr8_-_54091980 | 3.71 |
ENSMUST00000047768.11
|
Neil3
|
nei like 3 (E. coli) |
| chr7_-_127593003 | 3.68 |
ENSMUST00000033056.5
|
Pycard
|
PYD and CARD domain containing |
| chr3_-_51304137 | 3.50 |
ENSMUST00000141156.3
|
Mgarp
|
mitochondria localized glutamic acid rich protein |
| chr3_+_40905216 | 3.48 |
ENSMUST00000191872.6
ENSMUST00000200432.2 |
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr8_-_22675773 | 3.43 |
ENSMUST00000046916.9
|
Ckap2
|
cytoskeleton associated protein 2 |
| chr11_-_116357047 | 3.41 |
ENSMUST00000208602.2
|
Qrich2
|
glutamine rich 2 |
| chr11_-_73029070 | 3.34 |
ENSMUST00000052140.3
|
Haspin
|
histone H3 associated protein kinase |
| chr5_+_108280668 | 3.34 |
ENSMUST00000047677.9
|
Ccdc18
|
coiled-coil domain containing 18 |
| chr14_-_31299275 | 3.32 |
ENSMUST00000112027.9
|
Colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr2_+_18059982 | 3.25 |
ENSMUST00000028076.15
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr5_+_3393893 | 3.24 |
ENSMUST00000165117.8
ENSMUST00000197385.2 |
Cdk6
|
cyclin-dependent kinase 6 |
| chr9_-_57552844 | 3.23 |
ENSMUST00000216979.2
ENSMUST00000034863.8 |
Csk
|
c-src tyrosine kinase |
| chr2_-_163592127 | 3.22 |
ENSMUST00000017841.4
|
Ada
|
adenosine deaminase |
| chr5_+_21577640 | 3.19 |
ENSMUST00000035799.6
|
Fgl2
|
fibrinogen-like protein 2 |
| chr2_-_101479846 | 3.13 |
ENSMUST00000078494.6
ENSMUST00000160722.8 ENSMUST00000160037.8 |
Rag1
Iftap
|
recombination activating 1 intraflagellar transport associated protein |
| chr1_+_74430575 | 3.10 |
ENSMUST00000027367.14
|
Ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr17_+_47904355 | 3.09 |
ENSMUST00000182209.8
|
Ccnd3
|
cyclin D3 |
| chr13_+_38335232 | 3.08 |
ENSMUST00000124830.3
|
Dsp
|
desmoplakin |
| chr9_-_64080161 | 3.06 |
ENSMUST00000176299.8
ENSMUST00000130127.8 ENSMUST00000176794.8 ENSMUST00000177045.8 |
Zwilch
|
zwilch kinetochore protein |
| chr6_+_123206802 | 3.06 |
ENSMUST00000112554.9
ENSMUST00000024118.11 ENSMUST00000117130.8 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr10_+_20223516 | 2.98 |
ENSMUST00000169712.3
ENSMUST00000217608.2 |
Mtfr2
|
mitochondrial fission regulator 2 |
| chr10_-_81121596 | 2.94 |
ENSMUST00000218484.2
|
Tjp3
|
tight junction protein 3 |
| chr11_-_40624200 | 2.90 |
ENSMUST00000020579.9
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr9_-_107971640 | 2.90 |
ENSMUST00000081309.13
ENSMUST00000191985.2 |
Apeh
|
acylpeptide hydrolase |
| chr15_+_55171138 | 2.85 |
ENSMUST00000023053.12
ENSMUST00000110217.10 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr14_-_67953035 | 2.85 |
ENSMUST00000163100.8
ENSMUST00000132705.8 ENSMUST00000124045.3 |
Cdca2
|
cell division cycle associated 2 |
| chr15_-_103356880 | 2.85 |
ENSMUST00000065978.9
|
Gtsf2
|
gametocyte specific factor 2 |
| chr16_-_18880821 | 2.84 |
ENSMUST00000200568.2
|
Iglc1
|
immunoglobulin lambda constant 1 |
| chr1_+_190795198 | 2.77 |
ENSMUST00000076952.12
ENSMUST00000139340.8 ENSMUST00000078259.8 |
Nsl1
|
NSL1, MIS12 kinetochore complex component |
| chr7_+_28140352 | 2.67 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr10_-_75670217 | 2.66 |
ENSMUST00000218500.2
|
Gstt2
|
glutathione S-transferase, theta 2 |
| chr2_-_74489763 | 2.61 |
ENSMUST00000173623.2
ENSMUST00000001867.13 |
Evx2
|
even-skipped homeobox 2 |
| chr11_+_95734419 | 2.58 |
ENSMUST00000107708.2
|
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr6_+_41248311 | 2.57 |
ENSMUST00000103281.3
|
Trbv29
|
T cell receptor beta, variable 29 |
| chrX_-_8059597 | 2.56 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr5_-_123859070 | 2.51 |
ENSMUST00000031376.12
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr9_-_71803354 | 2.51 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr11_+_95734028 | 2.50 |
ENSMUST00000107709.8
|
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr1_+_107456731 | 2.49 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr8_+_85598734 | 2.48 |
ENSMUST00000170296.2
ENSMUST00000136026.8 |
Syce2
|
synaptonemal complex central element protein 2 |
| chr9_-_75317233 | 2.42 |
ENSMUST00000049355.11
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr15_-_97618982 | 2.42 |
ENSMUST00000023105.5
|
Endou
|
endonuclease, polyU-specific |
| chr14_+_75368532 | 2.41 |
ENSMUST00000143539.8
ENSMUST00000134114.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr1_+_180468895 | 2.40 |
ENSMUST00000192725.6
|
Lin9
|
lin-9 homolog (C. elegans) |
| chr1_-_59134042 | 2.39 |
ENSMUST00000238601.2
ENSMUST00000238949.2 ENSMUST00000097080.4 |
C2cd6
|
C2 calcium dependent domain containing 6 |
| chr1_-_93729650 | 2.37 |
ENSMUST00000027503.14
|
Dtymk
|
deoxythymidylate kinase |
| chr17_+_47904441 | 2.33 |
ENSMUST00000182874.3
|
Ccnd3
|
cyclin D3 |
| chr15_-_97629209 | 2.30 |
ENSMUST00000100249.10
|
Endou
|
endonuclease, polyU-specific |
| chr11_+_32155483 | 2.27 |
ENSMUST00000121182.2
|
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr18_+_66591604 | 2.27 |
ENSMUST00000025399.9
ENSMUST00000237161.2 ENSMUST00000236933.2 |
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr1_-_93729562 | 2.27 |
ENSMUST00000112890.3
|
Dtymk
|
deoxythymidylate kinase |
| chr7_+_12568647 | 2.26 |
ENSMUST00000004614.15
|
Zfp110
|
zinc finger protein 110 |
| chr9_-_106438798 | 2.25 |
ENSMUST00010126732.2
ENSMUST00010126033.2 ENSMUST00010181659.1 ENSMUST00010126065.2 ENSMUST00010126032.3 ENSMUST00000062917.16 |
ENSMUSG00001074846.1
ENSMUSG00000118396.3
|
IQ motif containing F3 IQ motif containing F3 |
| chr7_+_28869770 | 2.24 |
ENSMUST00000033886.8
ENSMUST00000209019.2 ENSMUST00000208330.2 |
Ggn
|
gametogenetin |
| chr6_-_70383976 | 2.21 |
ENSMUST00000103393.2
|
Igkv6-15
|
immunoglobulin kappa variable 6-15 |
| chr4_-_41464816 | 2.21 |
ENSMUST00000108055.9
ENSMUST00000154535.8 ENSMUST00000030148.6 |
Kif24
|
kinesin family member 24 |
| chr11_-_109718845 | 2.21 |
ENSMUST00000020941.11
|
1700012B07Rik
|
RIKEN cDNA 1700012B07 gene |
| chr10_+_28544556 | 2.19 |
ENSMUST00000161345.2
|
Themis
|
thymocyte selection associated |
| chr14_+_75368939 | 2.18 |
ENSMUST00000125833.8
ENSMUST00000124499.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr9_-_63509699 | 2.18 |
ENSMUST00000171243.2
ENSMUST00000163982.8 ENSMUST00000163624.8 |
Iqch
|
IQ motif containing H |
| chr14_+_29730931 | 2.18 |
ENSMUST00000067620.12
|
Chdh
|
choline dehydrogenase |
| chr6_+_86605146 | 2.14 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr6_+_146751598 | 2.12 |
ENSMUST00000032433.9
|
Smco2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr16_-_55659194 | 2.12 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr9_-_63509747 | 2.12 |
ENSMUST00000080527.12
ENSMUST00000042322.11 |
Iqch
|
IQ motif containing H |
| chr5_+_107977965 | 2.09 |
ENSMUST00000072578.8
|
Ube2d2b
|
ubiquitin-conjugating enzyme E2D 2B |
| chr19_-_43376794 | 2.09 |
ENSMUST00000099428.5
|
Hpse2
|
heparanase 2 |
| chr5_-_123859153 | 2.06 |
ENSMUST00000196282.5
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr7_+_28140450 | 2.05 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr9_-_36637670 | 2.05 |
ENSMUST00000172702.9
ENSMUST00000172742.2 |
Chek1
|
checkpoint kinase 1 |
| chr13_-_63036096 | 2.01 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr1_+_6285082 | 2.01 |
ENSMUST00000160062.8
|
Rb1cc1
|
RB1-inducible coiled-coil 1 |
| chr17_-_35424281 | 2.00 |
ENSMUST00000025266.6
|
Lta
|
lymphotoxin A |
| chr2_-_3423646 | 1.99 |
ENSMUST00000064685.14
|
Meig1
|
meiosis expressed gene 1 |
| chr11_-_82882613 | 1.96 |
ENSMUST00000092840.11
ENSMUST00000038211.13 |
Slfn9
|
schlafen 9 |
| chr17_-_40519480 | 1.93 |
ENSMUST00000033585.7
|
Pgk2
|
phosphoglycerate kinase 2 |
| chr4_+_98284054 | 1.91 |
ENSMUST00000107033.8
ENSMUST00000107034.8 |
Patj
|
PATJ, crumbs cell polarity complex component |
| chr17_+_35201074 | 1.88 |
ENSMUST00000007266.14
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_-_28945051 | 1.86 |
ENSMUST00000189711.7
ENSMUST00000157048.4 |
Cfap77
|
cilia and flagella associated protein 77 |
| chr9_-_40915858 | 1.85 |
ENSMUST00000188848.8
ENSMUST00000034519.13 |
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr7_-_97387429 | 1.84 |
ENSMUST00000206389.2
|
Aqp11
|
aquaporin 11 |
| chr7_-_4661980 | 1.83 |
ENSMUST00000205374.2
ENSMUST00000064099.8 |
Ppp6r1
|
protein phosphatase 6, regulatory subunit 1 |
| chr2_+_70339175 | 1.82 |
ENSMUST00000134607.8
|
Erich2
|
glutamate rich 2 |
| chr11_-_109718860 | 1.81 |
ENSMUST00000106674.8
|
1700012B07Rik
|
RIKEN cDNA 1700012B07 gene |
| chr12_-_101049884 | 1.81 |
ENSMUST00000048305.10
ENSMUST00000163095.9 |
Ppp4r3a
|
protein phosphatase 4 regulatory subunit 3A |
| chr4_-_112632013 | 1.79 |
ENSMUST00000060327.4
|
Skint10
|
selection and upkeep of intraepithelial T cells 10 |
| chr2_+_70339157 | 1.79 |
ENSMUST00000100041.9
|
Erich2
|
glutamate rich 2 |
| chr5_+_31046670 | 1.78 |
ENSMUST00000200850.2
|
Agbl5
|
ATP/GTP binding protein-like 5 |
| chr6_-_113412708 | 1.77 |
ENSMUST00000032416.11
ENSMUST00000113089.8 |
Cidec
|
cell death-inducing DFFA-like effector c |
| chr19_+_46385321 | 1.76 |
ENSMUST00000039922.13
ENSMUST00000111867.9 ENSMUST00000120778.8 |
Sufu
|
SUFU negative regulator of hedgehog signaling |
| chr8_+_41128099 | 1.76 |
ENSMUST00000051614.5
|
Adam24
|
a disintegrin and metallopeptidase domain 24 (testase 1) |
| chr6_+_40619913 | 1.75 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr9_-_36637923 | 1.75 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
| chrX_+_91533555 | 1.74 |
ENSMUST00000096371.3
|
Gm5941
|
predicted gene 5941 |
| chr14_-_75368321 | 1.74 |
ENSMUST00000022574.5
|
Lrrc63
|
leucine rich repeat containing 63 |
| chr11_+_72205264 | 1.72 |
ENSMUST00000108504.2
|
Fbxo39
|
F-box protein 39 |
| chr18_+_87774402 | 1.72 |
ENSMUST00000091776.7
|
Gm5096
|
predicted gene 5096 |
| chr4_-_123217391 | 1.72 |
ENSMUST00000102640.2
|
Oxct2a
|
3-oxoacid CoA transferase 2A |
| chr10_-_127502541 | 1.71 |
ENSMUST00000026469.9
|
Nab2
|
Ngfi-A binding protein 2 |
| chr9_+_95739650 | 1.71 |
ENSMUST00000034980.9
|
Atr
|
ataxia telangiectasia and Rad3 related |
| chr15_+_8138757 | 1.71 |
ENSMUST00000163765.3
|
Nup155
|
nucleoporin 155 |
| chr7_+_28869629 | 1.70 |
ENSMUST00000098609.4
|
Ggn
|
gametogenetin |
| chr7_-_4535403 | 1.68 |
ENSMUST00000094897.5
|
Dnaaf3
|
dynein, axonemal assembly factor 3 |
| chr19_+_46385448 | 1.67 |
ENSMUST00000118440.3
|
Sufu
|
SUFU negative regulator of hedgehog signaling |
| chr3_-_131065658 | 1.66 |
ENSMUST00000029610.9
|
Hadh
|
hydroxyacyl-Coenzyme A dehydrogenase |
| chr9_+_66853343 | 1.63 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr5_+_137627431 | 1.63 |
ENSMUST00000176667.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr2_-_143853122 | 1.62 |
ENSMUST00000016072.12
ENSMUST00000037875.6 |
Rrbp1
|
ribosome binding protein 1 |
| chr19_-_8751795 | 1.62 |
ENSMUST00000010249.7
|
Tmem179b
|
transmembrane protein 179B |
| chr15_+_8138805 | 1.61 |
ENSMUST00000230017.2
|
Nup155
|
nucleoporin 155 |
| chr5_+_145104011 | 1.61 |
ENSMUST00000160629.8
ENSMUST00000070487.12 ENSMUST00000160422.8 ENSMUST00000162244.8 |
Cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr15_-_76501041 | 1.61 |
ENSMUST00000073428.7
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr5_-_72661776 | 1.59 |
ENSMUST00000175766.8
ENSMUST00000177290.8 ENSMUST00000176974.2 ENSMUST00000167460.9 |
Corin
|
corin, serine peptidase |
| chr5_-_34326753 | 1.59 |
ENSMUST00000202409.2
ENSMUST00000202638.4 ENSMUST00000042954.11 ENSMUST00000202541.2 |
Poln
Haus3
|
DNA polymerase N HAUS augmin-like complex, subunit 3 |
| chr1_-_64161415 | 1.59 |
ENSMUST00000135075.2
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr11_+_100751272 | 1.58 |
ENSMUST00000107357.4
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr1_+_132243849 | 1.57 |
ENSMUST00000072177.14
ENSMUST00000082125.6 |
Nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr3_+_137983250 | 1.57 |
ENSMUST00000004232.10
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr8_+_11606056 | 1.55 |
ENSMUST00000210740.2
ENSMUST00000210670.2 ENSMUST00000054399.6 ENSMUST00000209646.2 |
Ing1
|
inhibitor of growth family, member 1 |
| chr11_-_82719850 | 1.54 |
ENSMUST00000021036.13
ENSMUST00000074515.11 ENSMUST00000103218.3 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr11_+_32155415 | 1.54 |
ENSMUST00000039601.10
ENSMUST00000149043.3 |
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr6_+_65019793 | 1.52 |
ENSMUST00000204696.3
|
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr7_-_130748035 | 1.50 |
ENSMUST00000070980.4
|
4933402N03Rik
|
RIKEN cDNA 4933402N03 gene |
| chr4_-_103148051 | 1.50 |
ENSMUST00000030245.10
ENSMUST00000168664.8 ENSMUST00000097944.4 |
4921539E11Rik
|
RIKEN cDNA 4921539E11 gene |
| chr17_+_87943401 | 1.50 |
ENSMUST00000235125.2
ENSMUST00000053577.9 ENSMUST00000234009.2 |
Epcam
|
epithelial cell adhesion molecule |
| chr8_+_22460606 | 1.50 |
ENSMUST00000017193.2
|
Ccdc70
|
coiled-coil domain containing 70 |
| chr10_-_127502467 | 1.49 |
ENSMUST00000099157.4
|
Nab2
|
Ngfi-A binding protein 2 |
| chr3_+_104688363 | 1.47 |
ENSMUST00000002298.7
|
Ppm1j
|
protein phosphatase 1J |
| chr1_-_119924920 | 1.47 |
ENSMUST00000174370.8
ENSMUST00000174458.2 |
Cfap221
|
cilia and flagella associated protein 221 |
| chr12_+_113453131 | 1.46 |
ENSMUST00000063317.4
|
Adam6b
|
a disintegrin and metallopeptidase domain 6B |
| chr14_+_53836282 | 1.45 |
ENSMUST00000103655.3
|
Trav4-3
|
T cell receptor alpha variable 4-3 |
| chr8_-_95564881 | 1.44 |
ENSMUST00000034233.15
ENSMUST00000162538.9 |
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr6_-_5256225 | 1.43 |
ENSMUST00000125686.8
|
Pon3
|
paraoxonase 3 |
| chr7_-_44803859 | 1.43 |
ENSMUST00000210125.2
|
Aldh16a1
|
aldehyde dehydrogenase 16 family, member A1 |
| chr12_-_114398864 | 1.42 |
ENSMUST00000103489.2
|
Ighv6-6
|
immunoglobulin heavy variable 6-6 |
| chr18_-_80751327 | 1.42 |
ENSMUST00000236310.2
ENSMUST00000167977.8 ENSMUST00000035800.8 |
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| chr5_-_107073704 | 1.41 |
ENSMUST00000200249.2
ENSMUST00000112690.10 |
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog |
| chr4_-_140573081 | 1.41 |
ENSMUST00000026378.4
|
Padi1
|
peptidyl arginine deiminase, type I |
| chr3_-_124374723 | 1.39 |
ENSMUST00000180162.8
ENSMUST00000047110.14 ENSMUST00000178485.8 |
1700003H04Rik
|
RIKEN cDNA 1700003H04 gene |
| chr11_+_100751151 | 1.38 |
ENSMUST00000138083.8
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr9_-_107971729 | 1.38 |
ENSMUST00000193254.6
|
Apeh
|
acylpeptide hydrolase |
| chrX_+_149372903 | 1.38 |
ENSMUST00000080884.11
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr9_+_95739747 | 1.37 |
ENSMUST00000215311.2
|
Atr
|
ataxia telangiectasia and Rad3 related |
| chr16_-_64299511 | 1.35 |
ENSMUST00000089279.5
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr6_+_65019574 | 1.34 |
ENSMUST00000031984.9
ENSMUST00000205118.3 |
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr7_-_26865838 | 1.33 |
ENSMUST00000108382.2
|
Egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr16_+_35758836 | 1.33 |
ENSMUST00000114878.8
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr8_+_88290469 | 1.33 |
ENSMUST00000093342.6
|
4933402J07Rik
|
RIKEN cDNA 4933402J07 gene |
| chr5_+_137627376 | 1.32 |
ENSMUST00000031734.16
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr14_+_54455293 | 1.31 |
ENSMUST00000103738.2
|
Traj2
|
T cell receptor alpha joining 2 |
| chr13_-_64236994 | 1.29 |
ENSMUST00000039832.7
|
Hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr4_+_122889407 | 1.27 |
ENSMUST00000144998.2
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
| chr9_+_106158549 | 1.26 |
ENSMUST00000191434.2
|
Poc1a
|
POC1 centriolar protein A |
| chr11_-_117764258 | 1.25 |
ENSMUST00000033230.8
|
Tha1
|
threonine aldolase 1 |
| chr15_+_81469538 | 1.24 |
ENSMUST00000068387.11
|
Ep300
|
E1A binding protein p300 |
| chrX_-_56438380 | 1.24 |
ENSMUST00000143310.2
ENSMUST00000098470.9 ENSMUST00000114726.8 |
Rbmx
|
RNA binding motif protein, X chromosome |
| chr6_-_5256282 | 1.23 |
ENSMUST00000031773.9
|
Pon3
|
paraoxonase 3 |
| chrX_+_100317629 | 1.22 |
ENSMUST00000117706.8
|
Med12
|
mediator complex subunit 12 |
| chr14_+_54082691 | 1.22 |
ENSMUST00000103674.6
|
Trav19
|
T cell receptor alpha variable 19 |
| chr1_+_88093726 | 1.20 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr2_-_151822114 | 1.20 |
ENSMUST00000062047.6
|
Fam110a
|
family with sequence similarity 110, member A |
| chr1_+_45350698 | 1.20 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr19_-_10460238 | 1.19 |
ENSMUST00000235392.2
ENSMUST00000237522.2 ENSMUST00000038842.5 |
Ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chrX_+_159173680 | 1.18 |
ENSMUST00000112408.9
ENSMUST00000112402.8 ENSMUST00000112401.8 ENSMUST00000112400.8 ENSMUST00000112405.9 ENSMUST00000112404.9 ENSMUST00000146805.8 |
Adgrg2
|
adhesion G protein-coupled receptor G2 |
| chr6_+_140569197 | 1.18 |
ENSMUST00000087614.11
|
Aebp2
|
AE binding protein 2 |
| chr8_+_123912976 | 1.16 |
ENSMUST00000019422.6
|
Dpep1
|
dipeptidase 1 |
| chr8_-_15096046 | 1.16 |
ENSMUST00000050493.4
ENSMUST00000123331.2 |
BB014433
|
expressed sequence BB014433 |
| chr5_-_121641461 | 1.14 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b |
| chr16_+_22769822 | 1.14 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr2_-_3423613 | 1.14 |
ENSMUST00000144584.2
|
Meig1
|
meiosis expressed gene 1 |
| chr11_+_74540284 | 1.14 |
ENSMUST00000117818.2
ENSMUST00000092915.12 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr4_+_137944546 | 1.14 |
ENSMUST00000130071.2
|
Hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr6_-_48817675 | 1.14 |
ENSMUST00000203265.3
ENSMUST00000205159.3 |
Tmem176b
|
transmembrane protein 176B |
| chr13_+_81805941 | 1.13 |
ENSMUST00000049055.8
|
Lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr14_-_99337137 | 1.13 |
ENSMUST00000042471.11
|
Dis3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease |
| chr14_+_54417419 | 1.13 |
ENSMUST00000103703.2
|
Traj39
|
T cell receptor alpha joining 39 |
| chr19_+_4175934 | 1.12 |
ENSMUST00000169055.10
ENSMUST00000135257.11 ENSMUST00000138593.10 ENSMUST00000140267.10 ENSMUST00000137579.6 ENSMUST00000139987.10 ENSMUST00000150627.11 ENSMUST00000151401.11 ENSMUST00000153654.11 |
Tmem134
|
transmembrane protein 134 |
| chr19_-_5538370 | 1.10 |
ENSMUST00000124334.8
|
Mus81
|
MUS81 structure-specific endonuclease subunit |
| chr6_-_88495835 | 1.10 |
ENSMUST00000032168.7
|
Sec61a1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr2_-_38895586 | 1.10 |
ENSMUST00000080861.6
|
Rpl35
|
ribosomal protein L35 |
| chr16_+_62674661 | 1.10 |
ENSMUST00000023629.9
|
Pros1
|
protein S (alpha) |
| chr9_-_44255456 | 1.10 |
ENSMUST00000077353.15
|
Hmbs
|
hydroxymethylbilane synthase |
| chr7_-_118455477 | 1.09 |
ENSMUST00000106550.11
|
Knop1
|
lysine rich nucleolar protein 1 |
| chr17_+_34544632 | 1.09 |
ENSMUST00000050325.10
|
H2-Eb2
|
histocompatibility 2, class II antigen E beta2 |
| chr1_-_139487951 | 1.07 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr6_+_52691204 | 1.07 |
ENSMUST00000138040.8
ENSMUST00000129660.2 |
Tax1bp1
|
Tax1 (human T cell leukemia virus type I) binding protein 1 |
| chr3_-_94922727 | 1.06 |
ENSMUST00000132195.8
|
Zfp687
|
zinc finger protein 687 |
| chr18_+_84869883 | 1.05 |
ENSMUST00000163083.2
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 1.6 | 4.7 | GO:0071846 | actin filament debranching(GO:0071846) |
| 1.5 | 4.6 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 1.4 | 9.5 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 1.3 | 3.8 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 1.2 | 3.7 | GO:0002582 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 1.1 | 3.3 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 1.1 | 3.2 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 1.1 | 3.2 | GO:0006154 | germinal center B cell differentiation(GO:0002314) adenosine catabolic process(GO:0006154) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) purine deoxyribonucleoside metabolic process(GO:0046122) negative regulation of mucus secretion(GO:0070256) |
| 1.0 | 3.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 1.0 | 3.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.9 | 2.7 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.8 | 3.3 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.8 | 3.1 | GO:1904868 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.8 | 4.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.7 | 3.0 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.7 | 2.0 | GO:0061723 | glycophagy(GO:0061723) |
| 0.7 | 2.7 | GO:0046226 | coumarin catabolic process(GO:0046226) |
| 0.7 | 2.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.7 | 3.3 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.5 | 1.0 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.5 | 2.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.5 | 3.4 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.5 | 1.4 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.5 | 1.8 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.4 | 3.5 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.4 | 1.7 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.4 | 2.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.4 | 6.7 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.4 | 1.2 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) |
| 0.4 | 1.6 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.4 | 1.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.4 | 1.5 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 1.1 | GO:0097037 | heme export(GO:0097037) |
| 0.4 | 1.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.4 | 1.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.4 | 2.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.3 | 1.0 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.3 | 1.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.3 | 3.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.3 | 3.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.3 | 1.0 | GO:1903978 | regulation of microglial cell activation(GO:1903978) |
| 0.3 | 2.3 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.3 | 2.3 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.3 | 1.6 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.3 | 1.5 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 | 1.2 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.3 | 0.9 | GO:1903413 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.3 | 1.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.3 | 1.4 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 1.8 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.3 | 0.8 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.3 | 1.3 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.2 | 11.6 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.2 | 0.7 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.2 | 1.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 3.7 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.2 | 3.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 2.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 0.7 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.2 | 5.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 3.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.2 | 0.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 1.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 2.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 4.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 2.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 1.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.2 | 0.5 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 1.8 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.2 | 1.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 1.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 2.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.2 | 2.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.2 | 0.9 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 1.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 10.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 1.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 1.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.9 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 1.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.6 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.7 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 1.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.8 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.7 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 3.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.2 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 3.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 1.3 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.1 | 0.6 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.5 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 1.0 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 1.3 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.8 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 1.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.9 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 2.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.8 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 3.1 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 0.3 | GO:0002856 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.1 | 0.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 1.3 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.1 | 2.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 3.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 1.0 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 0.6 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.9 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 0.9 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 3.1 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.1 | 0.9 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 3.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 1.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 3.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.6 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 1.2 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 4.3 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 0.4 | GO:2000049 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.1 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) sulfur oxidation(GO:0019417) |
| 0.1 | 1.8 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 1.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.8 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 1.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 3.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 3.4 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.7 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 1.7 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.8 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.0 | 6.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.7 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 3.0 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 2.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 1.0 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 2.8 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 2.6 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 1.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.4 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 1.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.5 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.1 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 0.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 1.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 1.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 1.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.6 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.7 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.5 | GO:0060044 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.1 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.1 | GO:0031392 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.0 | 0.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 2.2 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.4 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.4 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 1.7 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.6 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 1.7 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 7.6 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 1.6 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.7 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 2.4 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.0 | 0.1 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.0 | 1.4 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.4 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 1.1 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 2.2 | GO:0035107 | appendage morphogenesis(GO:0035107) limb morphogenesis(GO:0035108) |
| 0.0 | 0.3 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.4 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 1.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.8 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.5 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 3.1 | GO:0051321 | meiotic cell cycle(GO:0051321) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 16.5 | GO:0005818 | aster(GO:0005818) |
| 1.1 | 3.2 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 1.0 | 3.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.9 | 3.7 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.8 | 2.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.6 | 6.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.5 | 4.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.5 | 3.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.5 | 1.4 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.4 | 2.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.4 | 1.1 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.3 | 1.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.3 | 2.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.3 | 0.8 | GO:0005715 | chiasma(GO:0005712) late recombination nodule(GO:0005715) |
| 0.3 | 1.8 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.3 | 5.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 1.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 2.5 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 1.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 1.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 2.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 1.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 1.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 4.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 0.6 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.2 | 0.5 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 3.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 10.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.8 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 4.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 3.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.4 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 1.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 8.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 1.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 3.1 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 2.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.6 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.0 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 1.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 4.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 4.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.0 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 3.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 3.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 2.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 5.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 7.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 3.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 4.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.3 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 8.2 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 1.3 | 11.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 1.2 | 3.7 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 1.0 | 3.8 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.0 | 3.8 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.8 | 3.3 | GO:0072354 | histone kinase activity (H3-T3 specific)(GO:0072354) |
| 0.8 | 3.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.7 | 4.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.7 | 2.7 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.5 | 3.7 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.5 | 1.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.5 | 3.9 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.4 | 1.7 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.4 | 1.9 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.4 | 9.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.4 | 1.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.4 | 2.6 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.3 | 3.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.3 | 1.0 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.3 | 2.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.3 | 4.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 1.3 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.3 | 3.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 1.4 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.3 | 2.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.3 | 1.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 4.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 1.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 3.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.2 | 1.5 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.2 | 3.4 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.2 | 0.8 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 1.7 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.2 | 0.7 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.7 | GO:0071209 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.2 | 0.5 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 1.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.2 | 2.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 0.7 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.2 | 0.9 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.2 | 1.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.2 | 0.6 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.2 | 0.5 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 1.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 2.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.9 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 5.1 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 1.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 1.0 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 1.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.8 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 6.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 2.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 1.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.4 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 1.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.5 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.1 | 0.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 2.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 2.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 2.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 3.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.3 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 2.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 6.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 1.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.8 | GO:0001727 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.7 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 5.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 7.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 3.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 1.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 1.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 2.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 4.4 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.4 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 3.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 3.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 1.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 4.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 4.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 1.6 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 4.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 11.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 20.8 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 13.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 5.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 6.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 6.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 3.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 6.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 3.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 2.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 3.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 16.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.5 | 6.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 1.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 2.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 3.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 4.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 8.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 3.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 5.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 3.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 2.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 4.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 1.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 5.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 3.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 2.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 3.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 2.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |